2Q7V
 
 | | Crystal Structure of Deinococcus Radiodurans Thioredoxin Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Sanders, D.A.R, Obiero, J, Bonderoff, S.A, Goertzen, M.M. | | Deposit date: | 2007-06-07 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thioredoxin system from Deinococcus radiodurans.
J.Bacteriol., 192, 2010
|
|
1I8T
 
 | | STRUCTURE OF UDP-GALACTOPYRANOSE MUTASE FROM E.COLI | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Sanders, D.A.R, Staines, A.G, McMahon, S.A, McNeil, M.R, Whitfield, C, Naismith, J.H. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UDP-galactopyranose mutase has a novel structure and mechanism.
Nat.Struct.Biol., 8, 2001
|
|
1I3H
 
 | | CONCANAVALIN A-DIMANNOSE STRUCTURE | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Sanders, D.A.R, Moothoo, D.N, Raftery, J, Howard, A.J, Helliwell, J.R, Naismith, J.H. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution structure of the Con A-dimannose complex.
J.Mol.Biol., 310, 2001
|
|
3NTR
 
 | | Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6503 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3E4D
 
 | | Structural and Kinetic Study of an S-Formylglutathione Hydrolase from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, Esterase D, MAGNESIUM ION | | Authors: | Van Straaten, K.E, Gonzalez, C.F, Valladares, R.B, Xu, X, Savchenko, A.V, Sanders, D.A.R. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens
Protein Sci., 18, 2009
|
|
3HE3
 
 | |
3HDY
 
 | | Crystal Structure of UDP-galactopyranose mutase (reduced form) in complex with substrate | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Partha, S.K, van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate binding to UDP-galactopyranose mutase: crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP.
J.Mol.Biol., 394, 2009
|
|
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
6TZU
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
4RPJ
 
 | |
4RPK
 
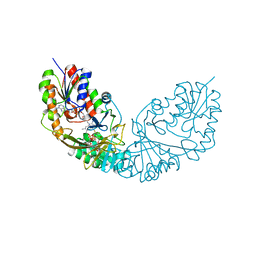 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galf | | Descriptor: | (2R,5S)-5-[(1R)-1,2-dihydroxyethyl]-3,3,4,4-tetrafluorotetrahydrofuran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4RPH
 
 | |
4RPL
 
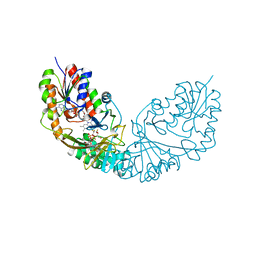 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,5S,6R)-3,3,4,4-tetrafluoro-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2499 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4RPG
 
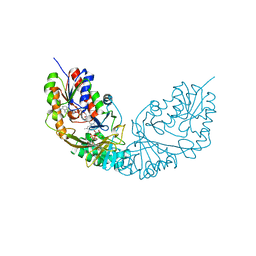 | |
5F1V
 
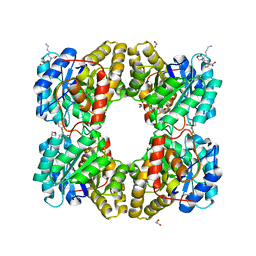 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
5F1U
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
6MOS
 
 | |
7LBD
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site in C2221 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Yazdi, M.M, Sanders, D.A.R. | | Deposit date: | 2021-01-07 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To Be Published
|
|
7LCF
 
 | |
7M06
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, Y110F mutant with R,R-bislysine bound at the allosteric site at 2.7 Angstrom | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Sanders, D.A.R. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | B-FACTOR ANALYSIS SUGGEST THAT L-LYSINE AND R, R-BISLYSINE ALLOSTERICALLY INHIBIT Cj.DHDPS ENZYME BY DECREASING PROTEIN DYNAMICS
To Be Published
|
|
4MO2
 
 | | Crystal Structure of UDP-N-acetylgalactopyranose mutase from Campylobacter jejuni | | Descriptor: | AMMONIUM ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Protsko, C, Poulin, M.B, Lowary, T.L, Sanders, D.A.R. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of a UDP-GalNAc Pyranose-Furanose Mutase: A Potential Therapeutic Target for Campylobacter jejuni Infections.
Chembiochem, 15, 2014
|
|
3MJ4
 
 | | Crystal structure of UDP-galactopyranose mutase in complex with phosphonate analog of UDP-galactopyranose | | Descriptor: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Karunan Partha, S, Sadeghi-Khomami, A, Slowski, K, Kotake, T, Thomas, N.R, Jakeman, D.L, Sanders, D.A.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemoenzymatic Synthesis, Inhibition Studies, and X-ray Crystallographic Analysis of the Phosphono Analog of UDP-Galp as an Inhibitor and Mechanistic Probe for UDP-Galactopyranose Mutase.
J.Mol.Biol., 403, 2010
|
|
3UKK
 
 | |
3MZ0
 
 | | Crystal structure of apo myo-inositol dehydrogenase from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-05-11 | | Release date: | 2010-09-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3UKQ
 
 | |
