4N6E
 
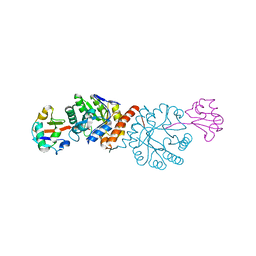 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | Descriptor: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y6K
 
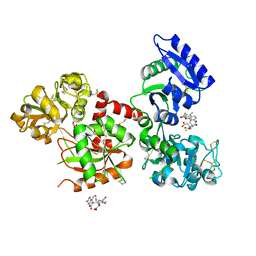 | | Human serum trnasferrin bound to a fluorescent probe | | Descriptor: | (2S)-6-[2-(7-azido-4-methyl-2-oxidanylidene-chromen-3-yl)ethanoylamino]-2-[bis(2-hydroxy-2-oxoethyl)amino]hexanoic acid, FE (III) ION, MALONATE ION, ... | | Authors: | Jiang, N, Cheng, T, Wang, M, Chan, G.C.F, Jin, L, Li, H, Sun, H. | | Deposit date: | 2017-08-12 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Tracking iron-associated proteomes in pathogens by a fluorescence approach.
Metallomics, 10, 2018
|
|
5Y2W
 
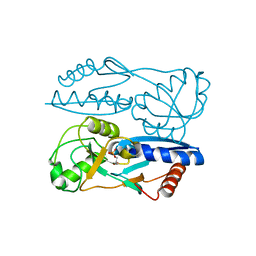 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4O0I
 
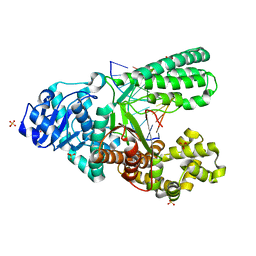 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with 2'-MeSe-arabino-guanosine derivatized DNA | | Descriptor: | 5'-D(*C*(1TW)P*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', DNA polymerase I, GLYCEROL, ... | | Authors: | Jiang, S, Gan, J, Sun, H, Huang, Z. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with 2'-MeSe-arabino-guanosine derivatized DNA
TO BE PUBLISHED
|
|
5Z49
 
 | | Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose-1,5-bisphosphate | | Descriptor: | HTH-type transcriptional activator CmpR, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Jiang, Y.L, Mahounga, D.M, Sun, H. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose 1,5-bisphosphate.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8W7E
 
 | | Design, synthesis and biological evaluations of novel small molecular hyper-activators of human caseinolytic peptidase P (hClpP) | | Descriptor: | 3-[(3-chlorophenyl)methyl]-6-[(4-chlorophenyl)methyl]-2,4-dihydro-1H-pyrido[2,3-c][2,7]naphthyridin-5-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Jiang, J.-X, Ding, H, Chen, M.-R, Lu, M.-L, Sun, H.-Y, Xiao, Y.-B. | | Deposit date: | 2023-08-30 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, synthesis and biological evaluations of novel small molecular hyper-activators of human caseinolytic peptidase P (hClpP)
To Be Published
|
|
8W7C
 
 | | Activation of mitochondrial Caseinolytic Protease P (ClpP) induces selective cancer cell lethality | | Descriptor: | 11-[(3-chlorophenyl)methyl]-7-[[4-(trifluoromethyl)phenyl]methyl]-2,5,7,11-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),3,5-trien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Jiang, J.-X, Ding, H, Lu, M.-L, Chen, M.-R, Sun, H.-Y, Xiao, Y.-B. | | Deposit date: | 2023-08-30 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation of mitochondrial Caseinolytic Protease P (ClpP) induces selective cancer cell lethality
To Be Published
|
|
8WUZ
 
 | | Development of 2-imino-2,3,5,6,7,8-hexahydropyrido[4,3-d]pyrimidin-4(1H)-one derivatives as human caseinolytic peptidase P (hClpP) activators | | Descriptor: | 5-[(3-fluorophenyl)methyl]-9-[[4-(trifluoromethyl)phenyl]methyl]-1,5,9,11-tetrazatricyclo[8.4.0.0^{2,7}]tetradeca-2(7),10-dien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Jiang, J.-X, Ding, H, Chen, M.-R, Lu, M.-L, Sun, H.-Y, Xiao, Y.-B. | | Deposit date: | 2023-10-22 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of 2-imino-2,3,5,6,7,8-hexahydropyrido[4,3-d]pyrimidin-4(1H)-one derivatives as human caseinolytic peptidase P (hClpP) activators
To Be Published
|
|
8W59
 
 | |
8W5A
 
 | |
7WAA
 
 | | Crystal structure of MCR-1-S treated by AgNO3 | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, SILVER ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Re-sensitization of mcr carrying multidrug resistant bacteria to colistin by silver.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
2H80
 
 | | NMR structures of SAM domain of Deleted in Liver Cancer 2 (DLC2) | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Li, H.Y, Fung, K.L, Jin, D.Y, Chung, S.S, Ko, B.C, Sun, H.Z. | | Deposit date: | 2006-06-06 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Proteins, 67, 2007
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
2F5H
 
 | | Solution structure of the alpha-domain of human Metallothionein-3 | | Descriptor: | CADMIUM ION, Metallothionein-3 | | Authors: | Wang, H, Zhang, Q, Cai, B, Li, H.Y, Sze, K.H, Huang, Z.X, Wu, H.M, Sun, H.Z. | | Deposit date: | 2005-11-25 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of human metallothionein-3 (MT-3)
Febs Lett., 580, 2006
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7CGC
 
 | |
7CGD
 
 | | Silver-bound E.coli malate dehydrogenase | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm.
Chem Sci, 11, 2020
|
|
7CB0
 
 | |
7CB5
 
 | |
