7K3X
 
 | |
7LUX
 
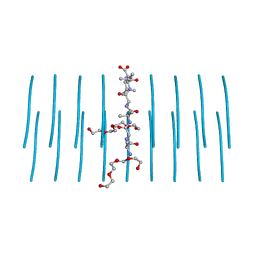 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7LUZ
 
 | |
7LV2
 
 | |
7LTU
 
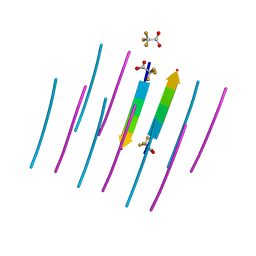 | | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2, RESIDUES 217-222, CRYSTAL FORM 1 | | Descriptor: | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2,RESIDUES 217-222, trifluoroacetic acid | | Authors: | Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.122 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7MGP
 
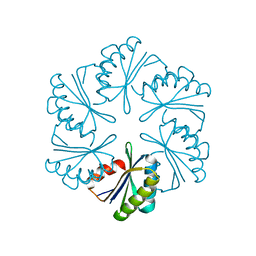 | | CmcA from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Escoto, X, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-04-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MPX
 
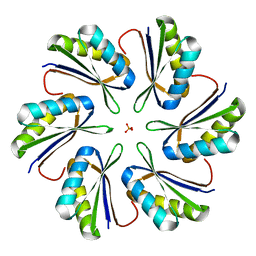 | | CmcC E36G mutant from Type II Cut MCP | | Descriptor: | BMC domain-containing protein, PHOSPHATE ION | | Authors: | Ochoa, J.M, Acosta, A.A, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MPW
 
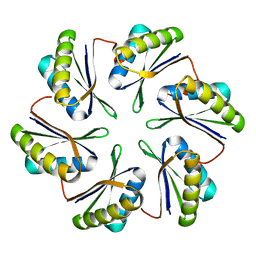 | | CmcB from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Marshall, J.D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MPV
 
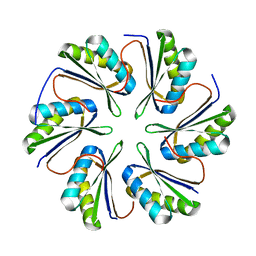 | | CmcC from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Mijares, O, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MN4
 
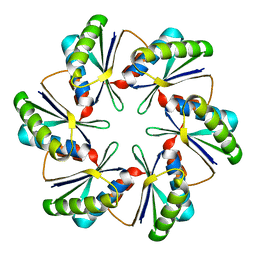 | | CmcB E35G mutant from Type II Cut MCP | | Descriptor: | BMC domain-containing protein | | Authors: | Ochoa, J.M, Mijares, O, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MMX
 
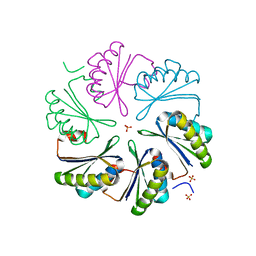 | | CutN from Type I Cut MCP | | Descriptor: | Propanediol utilization protein PduA, SULFATE ION | | Authors: | Ochoa, J.M, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7M62
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 2 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M65
 
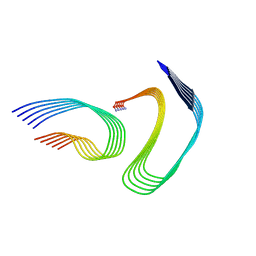 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 4 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M61
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 1 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M64
 
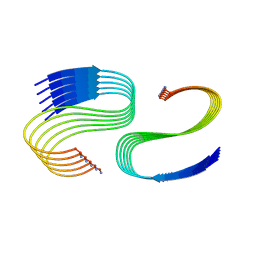 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 3 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
4NP8
 
 | | Structure of an amyloid forming peptide VQIVYK from the second repeat region of tau (alternate polymorph) | | Descriptor: | Microtubule-associated protein tau | | Authors: | Landau, M, Eisenberg, D, Sawaya, M.R, Dannenberg, J, Kobko, N. | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4OIK
 
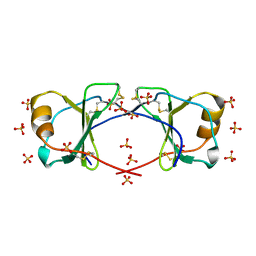 | | (Quasi-)Racemic X-ray crystal structure of glycosylated chemokine Ser-CCL1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1, CITRIC ACID, ... | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OIJ
 
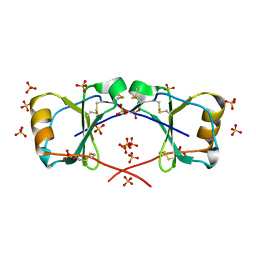 | | X-ray crystal structure of racemic non-glycosylated chemokine Ser-CCL1 | | Descriptor: | C-C motif chemokine 1, D-Ser-CCL1, SULFATE ION | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3L1E
 
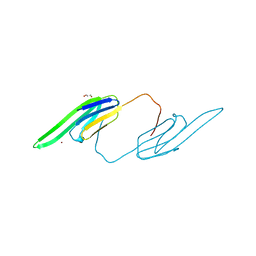 | | Bovine AlphaA crystallin Zinc Bound | | Descriptor: | Alpha-crystallin A chain, GLYCEROL, ZINC ION | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3L1G
 
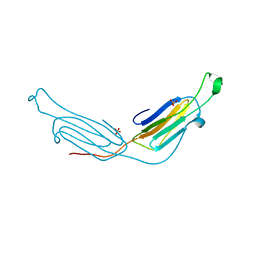 | | Human AlphaB crystallin | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
4ONK
 
 | | [Leu-5]-Enkephalin mutant - YVVFL | | Descriptor: | [Leu-5]-Enkephalin mutant - YVVFL | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
4OLP
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Pectobacterium wasabiae | | Descriptor: | GrpU microcompartment shell protein | | Authors: | Wheatley, N.M, Thompson, M.C, Gidaniyan, S.D, Sawaya, M.R, Jorda, J, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
4OLR
 
 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
4PPD
 
 | | PduA K26A, crystal form 2 | | Descriptor: | GLYCEROL, Propanediol utilization protein PduA, SULFATE ION | | Authors: | McNamara, D.E, Sawaya, M.R, Bobik, T.A, Yeates, T.O. | | Deposit date: | 2014-02-26 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the pdu microcompartment.
J.Mol.Biol., 426, 2014
|
|
4P2S
 
 | | Alanine Scanning Mutagenesis Identifies an Asparagine-Arginine-Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative propanediol utilization protein PduA, ... | | Authors: | Sinha, S, Cheng, S, Sung, Y.W, McNamara, D.E, Sawaya, M.R, Yeates, T.O, Bobik, T.A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alanine Scanning Mutagenesis Identifies an Asparagine-Arginine-Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment.
J.Mol.Biol., 426, 2014
|
|
