8G3K
 
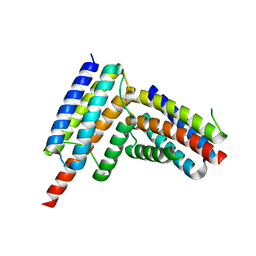 | |
8G47
 
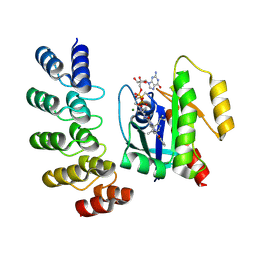 | | KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold | | Descriptor: | AMG 510 (bound form), GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4E
 
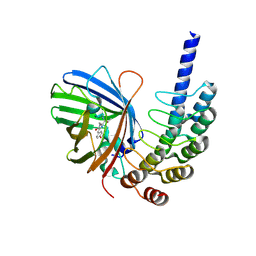 | |
8G4H
 
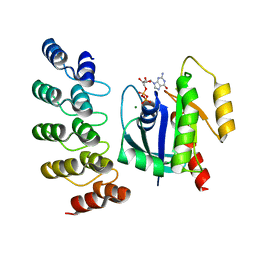 | | KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G42
 
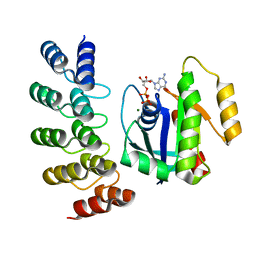 | | KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4F
 
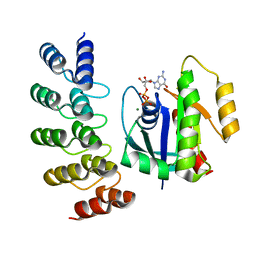 | | KRAS G12V complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G2V
 
 | | Cryo-EM structure of recombinant human LECT2 amyloid fibril core | | Descriptor: | Leukocyte cell-derived chemotaxin-2 | | Authors: | Richards, L.S, Flores, M.D, Zink, S, Schibrowsky, N.A, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.715 Å) | | Cite: | Cryo-EM structure of a human LECT2 amyloid fibril reveals a network of polar ladders at its core.
Structure, 31, 2023
|
|
4IQ4
 
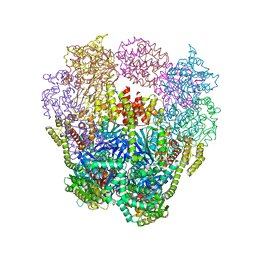 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P21212 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-10 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4ITV
 
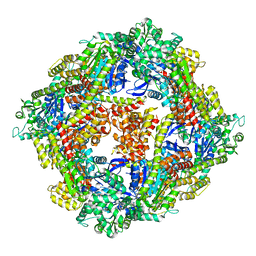 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P212121 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4K2X
 
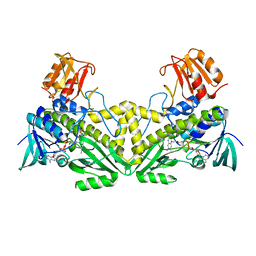 | |
4LCL
 
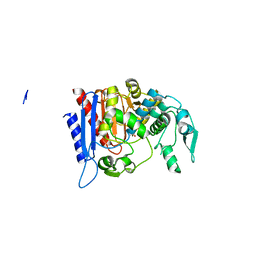 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD6 mutant (simh6208) | | Descriptor: | ISOPROPYL ALCOHOL, Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
4LCM
 
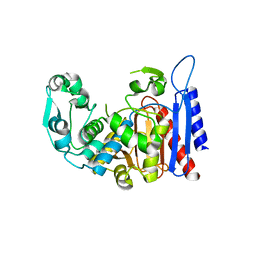 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD9 mutant (simh9014) | | Descriptor: | Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
4LFD
 
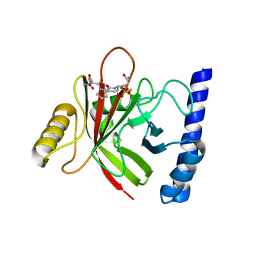 | | Staphylococcus aureus sortase B-substrate complex | | Descriptor: | (CBZ)NPQ(B27) PEPTIDE, SULFATE ION, Sortase B | | Authors: | Jacobitz, A.W, Sawaya, M.R, Yi, S.W, Amer, B.R, Huang, G.L, Nguyen, A.V, Jung, M.E, Clubb, R.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
J.Biol.Chem., 289, 2014
|
|
1KR5
 
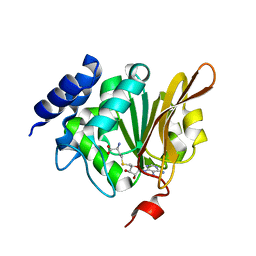 | | Crystal structure of human L-isoaspartyl methyltransferase | | Descriptor: | Protein-L-isoaspartate O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ryttersgaard, C, Griffith, S.C, Sawaya, M.R, MacLaren, D.C, Clarke, S, Yeates, T.O. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human L-isoaspartyl methyltransferase.
J.Biol.Chem., 277, 2002
|
|
1LMI
 
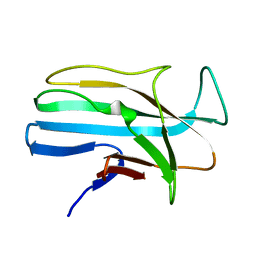 | | 1.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED PROTEIN FROM MYCOBACTERIUM TUBERCULOSIS-MPT63 | | Descriptor: | Immunogenic protein MPT63/MPB63 | | Authors: | Goulding, C.W, Parseghian, A, Sawaya, M.R, Cascio, D, Apostol, M, Gennaro, M.L, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-01 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at
1.5-A resolution
Protein Sci., 11, 2002
|
|
1MZ4
 
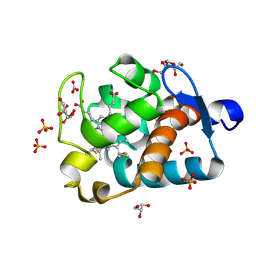 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
1I8F
 
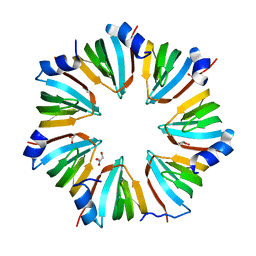 | | THE CRYSTAL STRUCTURE OF A HEPTAMERIC ARCHAEAL SM PROTEIN: IMPLICATIONS FOR THE EUKARYOTIC SNRNP CORE | | Descriptor: | GLYCEROL, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Mura, C, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2001-03-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1L9L
 
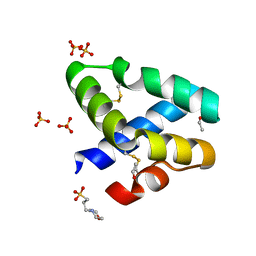 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1L6P
 
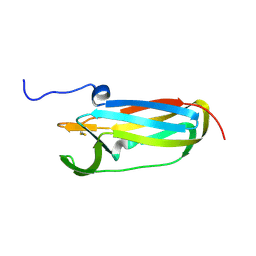 | |
1JG1
 
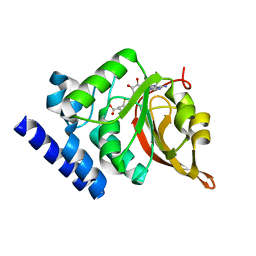 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
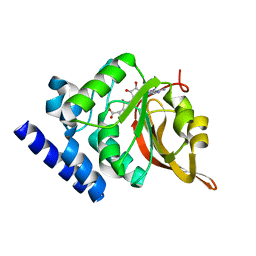 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1K26
 
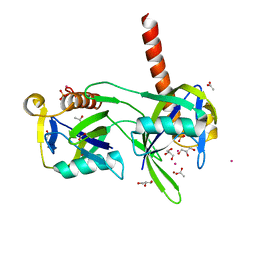 | | Structure of a Nudix Protein from Pyrobaculum aerophilum Solved by the Single Wavelength Anomolous Scattering Method | | Descriptor: | ACETIC ACID, GLYCEROL, IRIDIUM (III) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1NBU
 
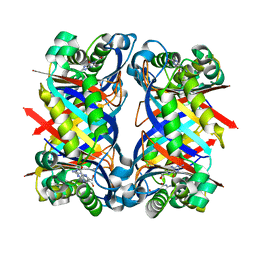 | | 7,8-Dihydroneopterin Aldolase Complexed with Product From Mycobacterium Tuberculosis | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, Probable dihydroneopterin aldolase | | Authors: | Goulding, C.W, Apostol, M.I, Sawaya, M.R, Phillips, M, Parseghian, A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-03 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme.
J.Mol.Biol., 349, 2005
|
|
1JG3
 
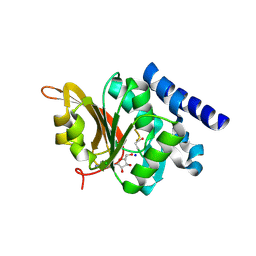 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1K2E
 
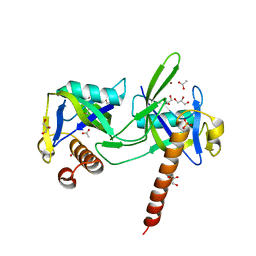 | | crystal structure of a nudix protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
