6MN2
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA
To Be Published
|
|
6BVC
 
 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
6MIJ
 
 | | Crystal structure of EF-Tu from Acinetobacter baumannii in complex with Mg2+ and GDP | | Descriptor: | Elongation factor Tu, FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | To be published
To Be Published
|
|
6NBK
 
 | | Crystal structure of Arginase from Bacillus cereus | | Descriptor: | Arginase, CALCIUM ION, MANGANESE (II) ION | | Authors: | Chang, C, Evdokimova, E, Mcchesney, M, Joachimiak, A, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Arginase from Bacillus cereus
To Be Published
|
|
6M8V
 
 | | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex
To Be Published
|
|
3NX3
 
 | | Crystal structure of acetylornithine aminotransferase (argD) from Campylobacter jejuni | | Descriptor: | Acetylornithine aminotransferase, MAGNESIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
5T07
 
 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Decanoyl-CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, decanoyl-CoA | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Decanoyl-CoA
To be published
|
|
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
5UM7
 
 | | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii | | Descriptor: | ACETATE ION, Thioredoxin signature protein | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-26 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii
To Be Published
|
|
5UC7
 
 | | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex
To Be Published
|
|
3IF4
 
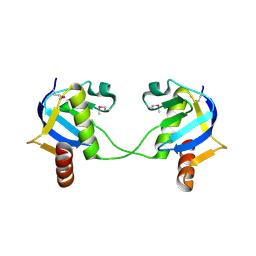 | | Structure from the mobile metagenome of North West Arm Sewage Outfall: Integron Cassette Protein Hfx_Cass5 | | Descriptor: | Integron Cassette Protein Hfx_Cass5 | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Evdokimova, E, Kudrytska, M, Koenig, J.E, Osipiuk, J, Edwards, A, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-24 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
3JRT
 
 | | Structure from the mobile metagenome of V. paracholerae: Integron cassette protein Vpc_cass2 | | Descriptor: | Integron cassette protein Vpc_cass2 | | Authors: | Harrop, S.J, Deshpande, C, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Cuff, M, Edwards, A, Savchenko, A, Joachimiak, A, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
3OP4
 
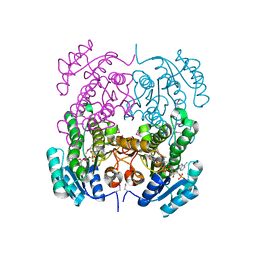 | | Crystal structure of putative 3-ketoacyl-(acyl-carrier-protein) reductase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with NADP+ | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Chruszcz, M, Onopriyenko, O, Grimshaw, S, Porebski, P, Zheng, H, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
5UF8
 
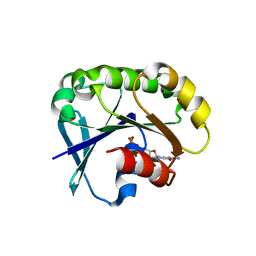 | | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Potential ADP-ribosylation factor | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP
To Be Published
|
|
4RHA
 
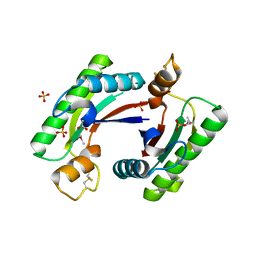 | | Structure of the C-terminal domain of outer-membrane protein OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Outer membrane protein A, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
5T06
 
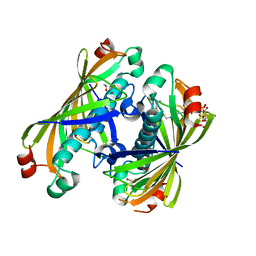 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase YbgC, HEXANOYL-COENZYME A | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA
To be published
|
|
3HJZ
 
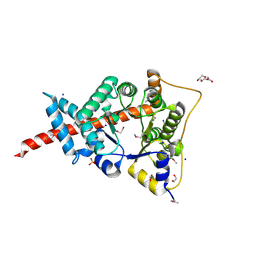 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1JW2
 
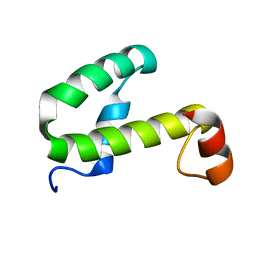 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5U8J
 
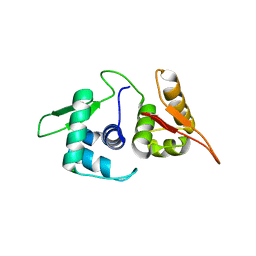 | | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae | | Descriptor: | CHLORIDE ION, UPF0502 protein BFJ73_07745 | | Authors: | Stogios, P.J, Skarina, T, Sandoval, J, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae
To Be Published
|
|
5UE7
 
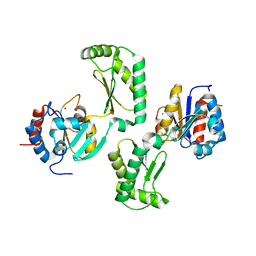 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
5V01
 
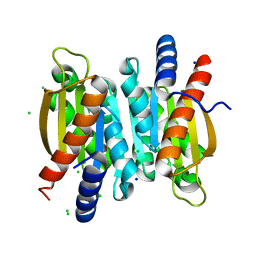 | | Crystal structure of the competence damage-inducible protein A (ComA) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | CHLORIDE ION, Competence damage-inducible protein A, SODIUM ION | | Authors: | Borek, D, Wawrzak, Z, Grimshaw, S, Sandoval, J, Evdokimova, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the competence damage-inducible protein A (ComA) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
5UCC
 
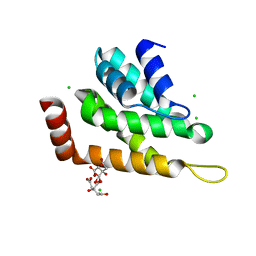 | | Crystal structure of the ENTH domain of ENT2 from Candida albicans | | Descriptor: | CHLORIDE ION, CITRIC ACID, Potential epsin-like clathrin-binding protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the ENTH domain of ENT2 from Candida albicans
To Be Published
|
|
3BJZ
 
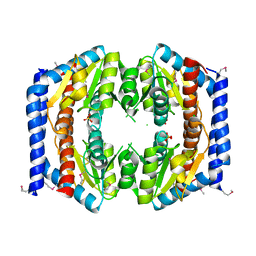 | | Crystal structure of Pseudomonas aeruginosa phosphoheptose isomerase | | Descriptor: | CHLORIDE ION, Phosphoheptose isomerase, SULFATE ION | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants.
J.Biol.Chem., 283, 2008
|
|
3MO4
 
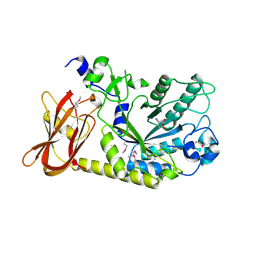 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
5UB8
 
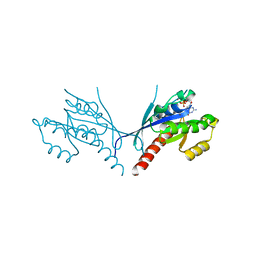 | | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Likely rab family GTP-binding protein, ZINC ION | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II)
To Be Published
|
|
