1YXO
 
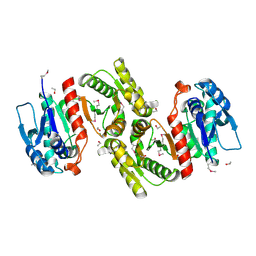 | | Crystal Structure of pyridoxal phosphate biosynthetic protein PdxA PA0593 | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, ETHANOL, MAGNESIUM ION | | Authors: | Liu, Y, Xu, X, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of pyridoxal phosphate biosynthetic protein PdxA PA0593
To be Published
|
|
1Z6N
 
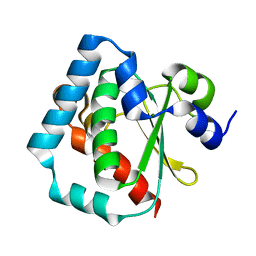 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA1234 from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, hypothetical protein PA1234 | | Authors: | Zhang, R, Xu, L, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1234 from Pseudomonas aeruginosa
To be Published
|
|
2FPO
 
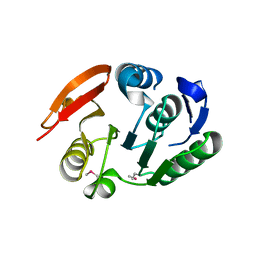 | | Putative methyltransferase yhhF from Escherichia coli. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, methylase yhhF | | Authors: | Osipiuk, J, Kim, Y, Sanishvili, R, Skarina, T, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-16 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure.
J.Biol.Chem., 282, 2007
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
2F2E
 
 | | Crystal Structure of PA1607, a Putative Transcription Factor | | Descriptor: | PA1607, SULFATE ION, alpha-D-glucopyranose | | Authors: | Sieminska, E.A, Xu, X, Zheng, H, Lunin, V, Cuff, M, Joachimiak, A, Edwards, A, Savchenko, A, Sanders, D.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-16 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The X-ray crystal structure of PA1607 from Pseudomonas aureginosa at 1.9 A resolution--a putative transcription factor.
Protein Sci., 16, 2007
|
|
3R1W
 
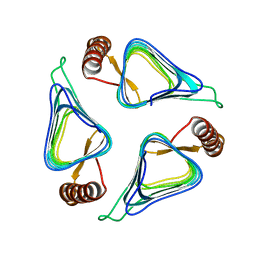 | | Crystal structure of a carbonic anhydrase from a crude oil degrading psychrophilic library | | Descriptor: | carbonic anhydrase | | Authors: | Petit, P, Xu, X, Cui, H, Brown, G, Dong, A, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a carbonic anhydrase from a crude oil degrading psychrophilic library
To be Published
|
|
2AS0
 
 | | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | | Descriptor: | hypothetical protein PH1915 | | Authors: | Sun, W, Xu, X, Pavlova, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
Protein Sci., 14, 2005
|
|
2G8Y
 
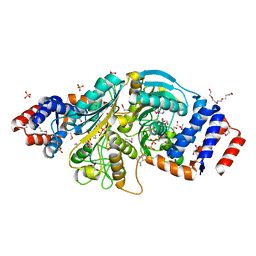 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
2G9I
 
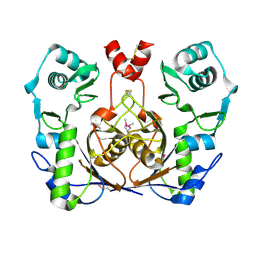 | | Crystal structure of homolog of F420-0:gamma-Glutamyl Ligase from Archaeoglobus fulgidus Reveals a Novel Fold. | | Descriptor: | F420-0:gamma-glutamyl ligase | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an amide bond forming F(420):gamma-glutamyl ligase from Archaeoglobus fulgidus -- a member of a new family of non-ribosomal peptide synthases.
J.Mol.Biol., 372, 2007
|
|
2GYQ
 
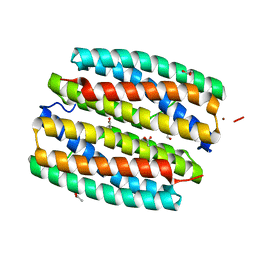 | | YcfI, a putative structural protein from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of YcfI protein, a putative structural protein from Rhodopseudomonas palustris.
To be Published
|
|
6ALL
 
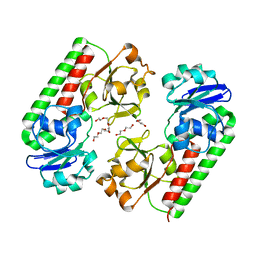 | | Crystal structure of a predicted ferric/iron (III) hydroxymate siderophore substrate binding protein from Bacillus anthracis | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Fe(3+)-citrate-binding protein yfmC | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of a predicted ferric/iron (III) hydroxymate siderophore substrate binding protein from Bacillus anthracis
To Be Published
|
|
3QVM
 
 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | Authors: | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
5TVL
 
 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
3SVI
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited thermolysin digestion | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | Descriptor: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
3RF6
 
 | | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, MAGNESIUM ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Nocek, B, Kuznetsova, K, Evdokimova, E, Savchenko, A, Iakunine, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-05 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae
TO BE PUBLISHED
|
|
3QVQ
 
 | | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Cuff, M.E, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate
To be Published
|
|
6M8U
 
 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, prenylated-FMN complex | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, prenylated-FMN complex
To Be Published
|
|
6M8T
 
 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidnova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex
To Be Published
|
|
6MR3
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans | | Descriptor: | CHLORIDE ION, Putative competence-damage inducible protein | | Authors: | Stogios, P.J, Cuff, M, Xu, X, Cui, H, Di Leo, R, Yim, V, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans
To Be Published
|
|
6NPS
 
 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | Descriptor: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | Deposit date: | 2019-01-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
3GOA
 
 | | Crystal structure of the Salmonella typhimurium FadA 3-ketoacyl-CoA thiolase | | Descriptor: | 3-ketoacyl-CoA thiolase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, S.M, Skarina, T, Onopriyenko, O, Wawrzak, Z, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: |
|
|
3QM1
 
 | | CRYSTAL STRUCTURE OF THE LACTOBACILLUS JOHNSONII CINNAMOYL ESTERASE LJ0536 S106A MUTANT IN COMPLEX WITH ETHYLFERULATE, Form II | | Descriptor: | CHLORIDE ION, Cinnamoyl esterase, SODIUM ION, ... | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2011-02-03 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
3DM8
 
 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
