4XTK
 
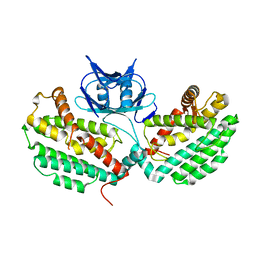 | | Structure of TM1797, a CAS1 protein from Thermotoga maritima | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Beloglazova, N, Skarina, T, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and nuclease activity of tm1797, a cas1 protein from thermotoga maritima
To Be Published
|
|
5JOQ
 
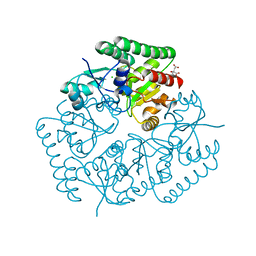 | | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, CITRIC ACID, Lmo2184 protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-02 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e
To Be Published
|
|
4WZ3
 
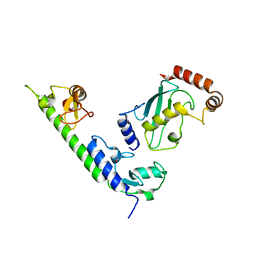 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4WZ2
 
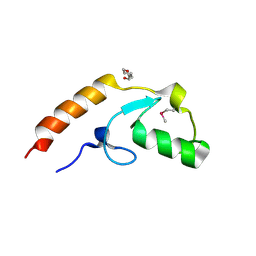 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | Authors: | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-28 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4WZ0
 
 | | Crystal structure of U-box 1 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris | | Descriptor: | E3 ubiquitin-protein ligase LubX | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Stein, A, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-14 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
5KVR
 
 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
5L0L
 
 | | Crystal structure of Uncharacterized protein LPG0439 | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Chang, C, Skarina, T, Khutoreskaya, G, Savchenko, A, Joachimiak, A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Uncharacterized protein LPG0439
To Be Published
|
|
5IQJ
 
 | | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae.
To Be Published
|
|
4XUV
 
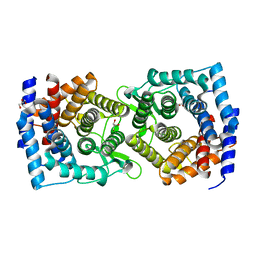 | | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris | | Descriptor: | GLYCEROL, Glycoside hydrolase family 105 protein | | Authors: | Stogios, P.J, Xu, X, Cui, H, Yim, V, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0496 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris
To Be Published
|
|
4XUY
 
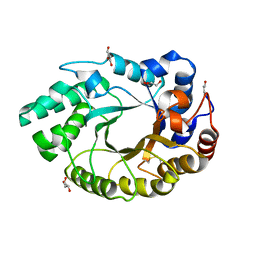 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger | | Descriptor: | GLYCEROL, Probable endo-1,4-beta-xylanase C, SULFATE ION | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0027 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger
To Be Published
|
|
1PVM
 
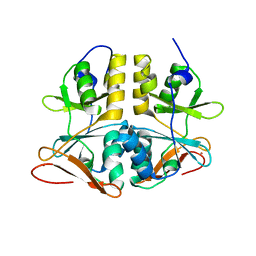 | | Crystal Structure of a Conserved CBS Domain Protein TA0289 of Unknown Function from Thermoplasma acidophilum | | Descriptor: | MERCURY (II) ION, conserved hypothetical protein Ta0289 | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Xu, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-27 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain
J.Mol.Biol., 375, 2008
|
|
4WRP
 
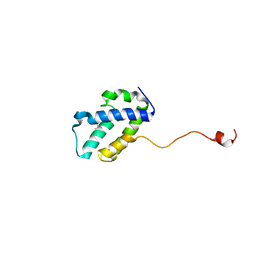 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
4WJ0
 
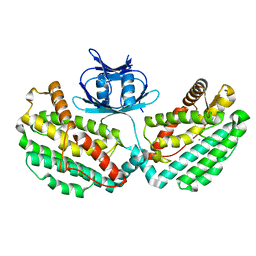 | | Structure of PH1245, a cas1 from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Brown, G, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of PH1245, a cas1 from Pyrococcus horikoshii
To Be Published
|
|
7UXG
 
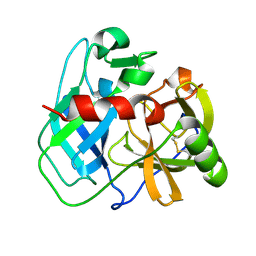 | | Crystal structure of putative serine protease YdgD from Escherichia coli | | Descriptor: | Serine protease | | Authors: | Stogios, P.J, Michalska, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of putative serine protease YdgD from Escherichia coli
To Be Published
|
|
4ZOS
 
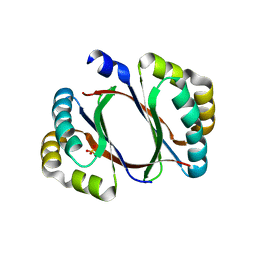 | | 2.20 Angstrom resolution crystal structure of protein YE0340 of unidentified function from Yersinia enterocolitica subsp. enterocolitica 8081] | | Descriptor: | PHOSPHATE ION, protein YE0340 from Yersinia enterocolitica subsp. enterocolitica 8081 | | Authors: | Halavaty, A.S, Wawrzak, A, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.20 Angstrom resolution crystal structure of protein YE0340 of unidentified function from Yersinia enterocolitica subsp. enterocolitica 8081]
To Be Published
|
|
4XCW
 
 | |
7T9W
 
 | | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2
To Be Published
|
|
7TI9
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-13 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2
To Be Published
|
|
5JD8
 
 | | Crystal structure of the serine endoprotease from Yersinia pestis | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DI(HYDROXYETHYL)ETHER, Periplasmic serine peptidase DegS, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the serine endoprotease from Yersinia pestis
To Be Published
|
|
7V09
 
 | | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein | | Descriptor: | MAGNESIUM ION, Multiple sugar transport system periplasmic sugar-binding protein | | Authors: | Stogios, P.J, Skarina, T, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein
To Be Published
|
|
3ICF
 
 | | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Cui, H, Kagan, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5
To be Published
|
|
3ICC
 
 | | Crystal structure of a putative 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis at 1.87 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cymborowski, M, Luo, H.-B, Skarina, T, Gordon, S, Savchenko, A, Edwards, A.M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a short-chain dehydrogenase/reductase from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3IV4
 
 | | A putative oxidoreductase with a thioredoxin fold | | Descriptor: | Putative oxidoreductase | | Authors: | Fan, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a putative oxidoreductase with a thioredoxin fold
To be Published
|
|
3I6Y
 
 | | Structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
3IG4
 
 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
