3N5U
 
 | | Crystal structure of an Rb C-terminal peptide bound to the catalytic subunit of PP1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Retinoblastoma-associated protein, ... | | Authors: | Hirschi, A.M, Cecchini, M, Steinhardt, R.C, Dick, F.A, Rubin, S.M. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NI0
 
 | | Crystal Structure of Mouse BST-2/Tetherin Ectodomain | | Descriptor: | Bone marrow stromal antigen 2, ISOPROPYL ALCOHOL | | Authors: | Scheaffer, S.M, Brett, T.J. | | Deposit date: | 2010-06-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biophysical Analysis of BST-2/Tetherin Ectodomains Reveals an Evolutionary Conserved Design to Inhibit Virus Release.
J.Biol.Chem., 286, 2011
|
|
3IDW
 
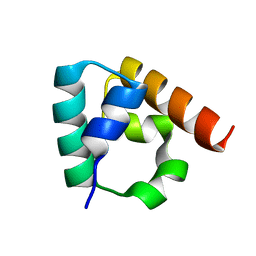 | |
3IXX
 
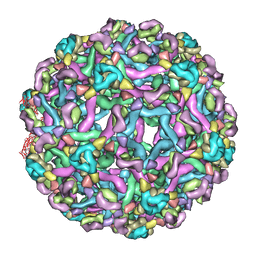 | | The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
1PA4
 
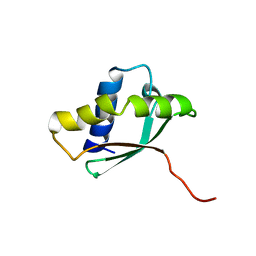 | | Solution structure of a putative ribosome-binding factor from Mycoplasma pneumoniae (MPN156) | | Descriptor: | Probable ribosome-binding factor A | | Authors: | Rubin, S.M, Pelton, J.G, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-05-13 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative ribosome binding protein from Mycoplasma pneumoniae and comparison to a distant homolog.
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1P7O
 
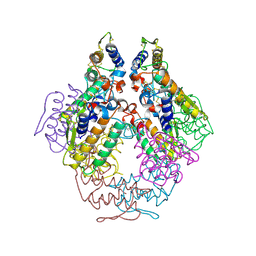 | |
4Q7V
 
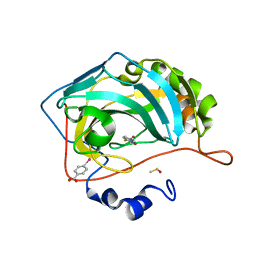 | |
4Q83
 
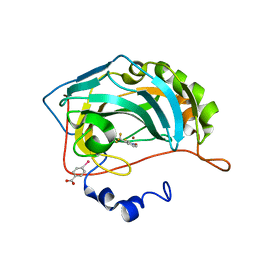 | |
4Q8X
 
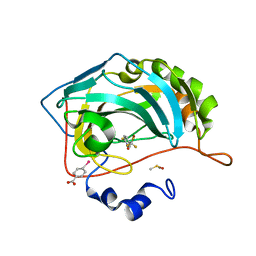 | |
4Q99
 
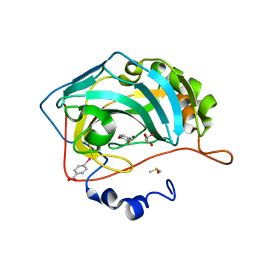 | |
1OZY
 
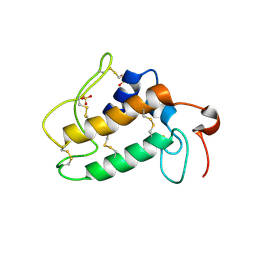 | |
4Q6A
 
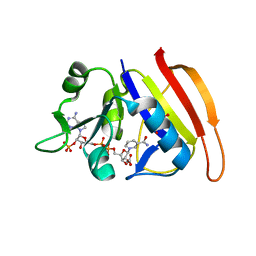 | |
3J05
 
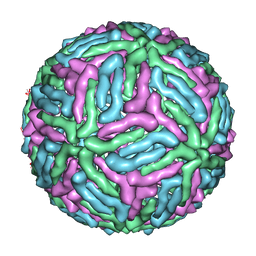 | | Three-dimensional structure of Dengue virus serotype 1 complexed with HMAb 14c10 Fab | | Descriptor: | envelope protein | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A, Tan, T.T, Shi, P.Y, Yip, A, Schul, W, Leo, Y.S, Chan, S.H, Smith, K.G.C, Ooi, E.E, Kemeny, D.M, Ng, G, Ng, M.L, Alonso, S, Fisher, D, Hanson, B, Lok, S.M, MacAry, P.A. | | Deposit date: | 2011-04-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The structural basis for serotype-specific neutralization of dengue virus by a human antibody.
Sci Transl Med, 4, 2012
|
|
3J7E
 
 | | Electron cryo-microscopy of human papillomavirus 16 and H16.V5 Fab fragments | | Descriptor: | H16.V5 Fab heavy chain, H16.V5 Fab light chain | | Authors: | Lee, H, Brendle, S.A, Bywaters, S.M, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2014-06-23 | | Release date: | 2014-11-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment.
J.Virol., 89, 2015
|
|
3J4K
 
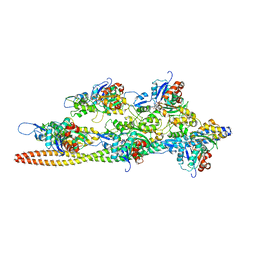 | | Cryo-EM structures of the actin:tropomyosin filament reveal the mechanism for the transition from C- to M-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sousa, D.R, Stagg, S.M, Stroupe, M.E. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM Structures of the Actin:Tropomyosin Filament Reveal the Mechanism for the Transition from C- to M-State.
J.Mol.Biol., 425, 2013
|
|
2P26
 
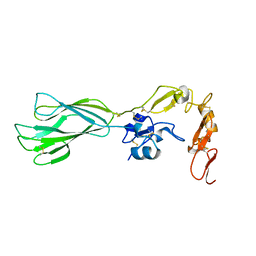 | | Structure of the PHE2 and PHE3 fragments of the integrin beta2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin beta-2 | | Authors: | Shi, M, Foo, S.Y, Tan, S.M, Mitchell, E.P, Law, S.K.A, Lescar, J. | | Deposit date: | 2007-03-06 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
J.Biol.Chem., 282, 2007
|
|
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
4M80
 
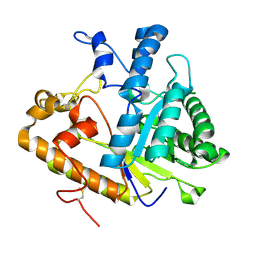 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution | | Descriptor: | EXO-1,3-BETA-GLUCANASE | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
4MDT
 
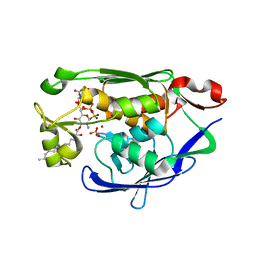 | | Structure of LpxC bound to the reaction product UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine | | Descriptor: | PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Clayton, G.M, Klein, D.J, Rickert, K.W, Patel, S.B, Kornienko, M, Zugay-Murphy, J, Reid, J.C, Tummala, S, Sharma, S, Singh, S.B, Miesel, L, Lumb, K.J, Soisson, S.M. | | Deposit date: | 2013-08-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer.
J.Biol.Chem., 288, 2013
|
|
4MAE
 
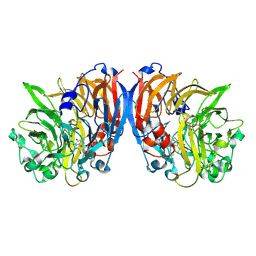 | | Methanol dehydrogenase from Methylacidiphilum fumariolicum SolV | | Descriptor: | CERIUM (III) ION, Methanol dehydrogenase, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Pol, A.J, Barends, T.R.M, Dietl, A, Khadem, A.F, Eygenstein, J, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2013-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare earth metals are essential for methanotrophic life in volcanic mudpots.
ENVIRON.MICROBIOL., 16, 2014
|
|
4LTD
 
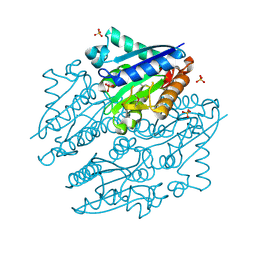 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - apo form | | Descriptor: | NADH-dependent FMN reductase, PHOSPHATE ION, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4M32
 
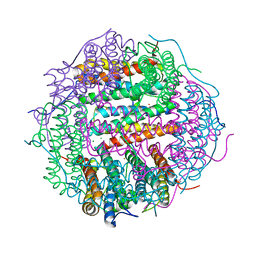 | | Crystal structure of gated-pore mutant D138N of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
2YEQ
 
 | | Structure of PhoD | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALKALINE PHOSPHATASE D, ... | | Authors: | Lillington, J.E.D, Rodriguez, F, Roversi, P, Johnson, S.J, Berks, B, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Phosphodiesterase Phod Reveals an Iron and Calcium-Containing Active Site.
J.Biol.Chem., 289, 2014
|
|
2PK7
 
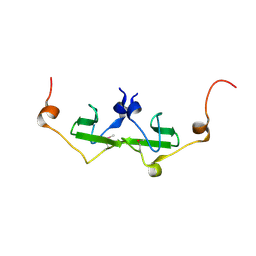 | | Crystal structure of the Q4KFT4_PSEF5 protein from Pseudomonas fluorescens. NESG target PlR1 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Neely, H, Jayaraman, S, Chen, C.X, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Q4KFT4_PSEF5 protein from Pseudomonas fluorescens.
To be Published
|
|
5FM9
 
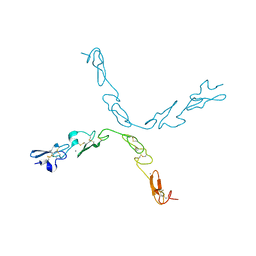 | | human Notch 1, EGF 4-7 | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Weisshuhn, P.C, Sheppard, D, Taylor, P, Whiteman, P, Lea, S.M, Handford, P.A, Redfield, C. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Non-Linear and Flexible Regions of the Human Notch1 Extracellular Domain Revealed by High-Resolution Structural Studies.
Structure, 24, 2016
|
|
