6R69
 
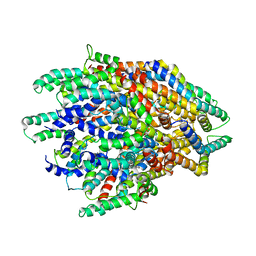 | | Improved map of the FliPQR complex that forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
6OM6
 
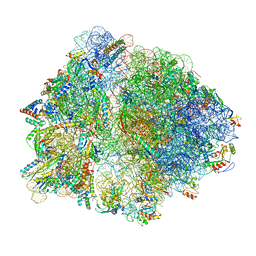 | | Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Mehrani, A, Keiler, K.C, Stagg, S.M, Dunham, C.M. | | Deposit date: | 2019-04-18 | | Release date: | 2021-02-10 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | trans-Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo.
Nat Commun, 12, 2021
|
|
7RJ2
 
 | |
7RJ4
 
 | |
6OSW
 
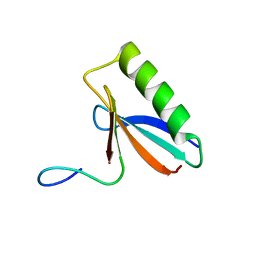 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
7RHN
 
 | |
7RHM
 
 | |
7RMJ
 
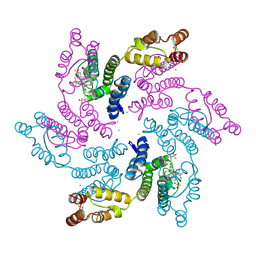 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor (S)-N-(1-(3-(4-chloro-3-(methylsulfonamido)-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl)-6-(3-methyl-3-(methylsulfonyl)but-1-yn-1-yl)pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CAPSID PROTEIN P24, CHLORIDE ION, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and Mechanistic Bases of Viral Resistance to HIV-1 Capsid Inhibitor Lenacapavir.
Mbio, 13, 2022
|
|
7RMM
 
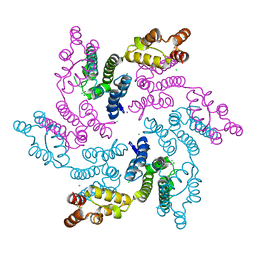 | |
7RAO
 
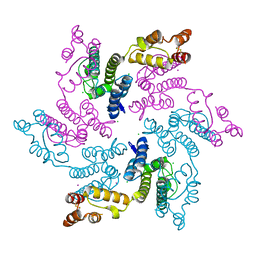 | |
6R6B
 
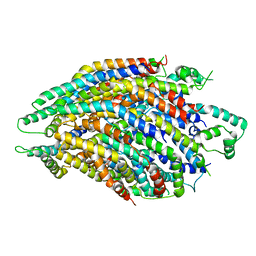 | | Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29). | | Descriptor: | Surface presentation of antigens protein SpaP, Surface presentation of antigens protein SpaQ, Surface presentation of antigens protein SpaR | | Authors: | Johnson, S, Kuhlen, L, Deme, J.C, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
8IZE
 
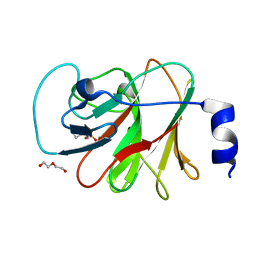 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IXV
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A in complex with 2Cl-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IZG
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 5-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JBQ
 
 | |
8JC7
 
 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
6RR0
 
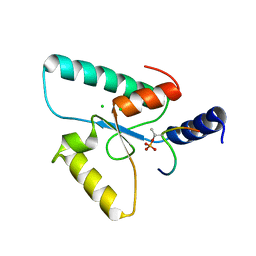 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ubp10 pT123 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6RU3
 
 | |
8JN7
 
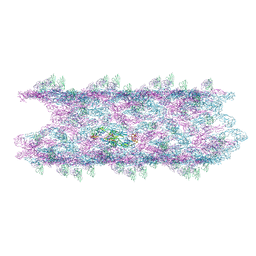 | | Cryo-EM structure of the big tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN3
 
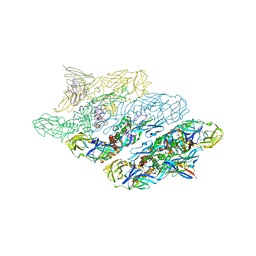 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C (subparticle LLR-LRR) | | Descriptor: | Envelope protein, Human antibody DENV-115 heavy chain, Human antibody DENV-115 light chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN6
 
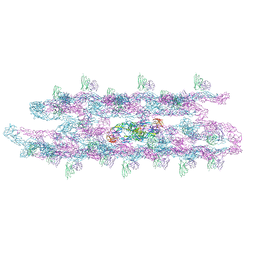 | | Cryo-EM structure of the small tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8OUO
 
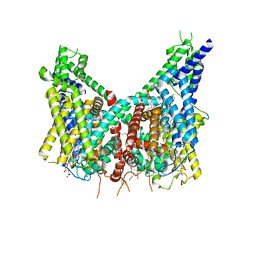 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 32, 2024
|
|
7V9R
 
 | | Crystal Structure of the heptameric EcHsp60 | | Descriptor: | 60 kDa chaperonin | | Authors: | Lai, M.C, Lin, S.M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of dimeric and heptameric mtHsp60 reveal the mechanism of chaperonin inactivation.
Life Sci Alliance, 6, 2023
|
|
7V98
 
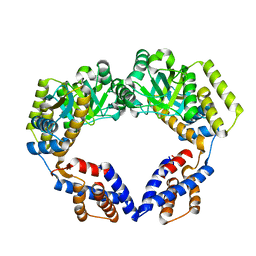 | | Crystal Structure of the Dimeric EcHsp60 | | Descriptor: | 60 kDa chaperonin | | Authors: | Lai, M.C, Lin, S.M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of dimeric and heptameric mtHsp60 reveal the mechanism of chaperonin inactivation.
Life Sci Alliance, 6, 2023
|
|
7NPO
 
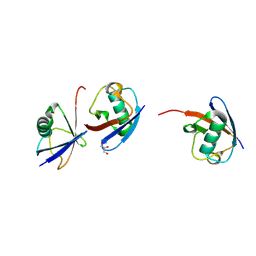 | | Branched K48-K63-Ub3 | | Descriptor: | GLYCEROL, Polyubiquitin-B | | Authors: | Lange, S.M, Kwasna, D, Kulathu, Y. | | Deposit date: | 2021-02-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VCP/p97-associated proteins are binders and debranching enzymes of K48-K63-branched ubiquitin chains.
Nat.Struct.Mol.Biol., 2024
|
|
