8UHG
 
 | |
4Q7V
 
 | |
4Q83
 
 | |
4Q8X
 
 | |
8UIS
 
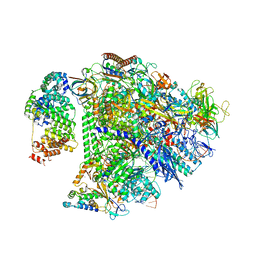 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8UHA
 
 | |
3GQV
 
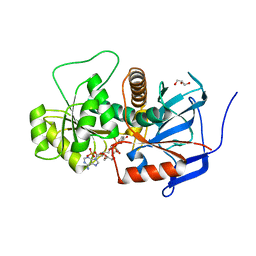 | | Lovastatin polyketide enoyl reductase (LovC) mutant K54S with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Kaake, R, Wong, E.W, Wong, S.K, Xie, X, Li, J.W, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | biosynthesis of Lovastatin: Crystal structure and biochemical
studies of LOVC, A trans-acting polyketide enoyl reductase
To be Published
|
|
8UHD
 
 | |
2BG7
 
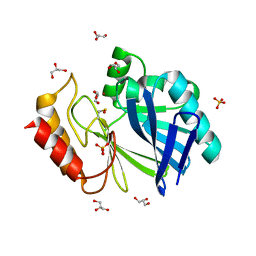 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
2F9C
 
 | | Crystal structure of YDCK from Salmonella cholerae. NESG target SCR6 | | Descriptor: | CESIUM ION, Hypothetical protein YDCK | | Authors: | Benach, J, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, H, Cooper, B, Acton, X.T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-05 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YDCK from Salmonella cholerae NESG target SCR6.
To be Published
|
|
3LS0
 
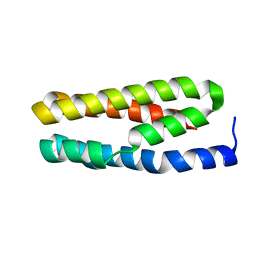 | |
2BGG
 
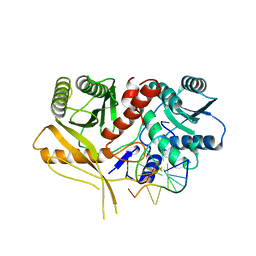 | | The structure of a Piwi protein from Archaeoglobus fulgidus complexed with a 16nt siRNA duplex. | | Descriptor: | 5'-R(*GP*UP*CP*GP*AP*AP*UP*UP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*GP*CP)-3', MANGANESE (II) ION, ... | | Authors: | Parker, J.S, Roe, S.M, Barford, D. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Mrna Recognition from a Piwi Domain-Sirna Guide Complex
Nature, 434, 2005
|
|
3HRU
 
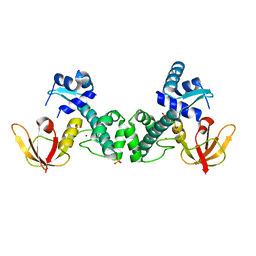 | | Crystal Structure of ScaR with bound Zn2+ | | Descriptor: | Metalloregulator ScaR, SULFATE ION, ZINC ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
4MLX
 
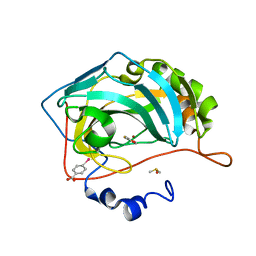 | |
2HDK
 
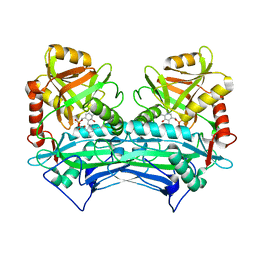 | |
4Q99
 
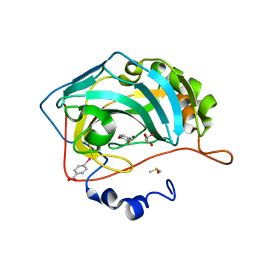 | |
2GWL
 
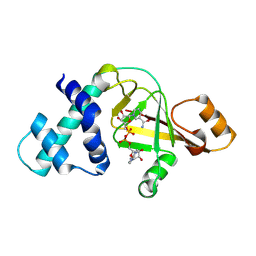 | |
1SKT
 
 | | SOLUTION STRUCTURE OF APO N-DOMAIN OF TROPONIN C, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
2BG2
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in the buffer. 1mM DTT and 1mM TCEP- HCl were used as reducing agents. Cys221 is reduced. | | Descriptor: | BETA-LACTAMASE II, CHLORIDE ION, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism.
Biochemistry, 44, 2005
|
|
2FFG
 
 | | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360. | | Descriptor: | ykuJ | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Ho, C.K, Janjua, H, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360.
To be Published
|
|
2BFZ
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
4Q6A
 
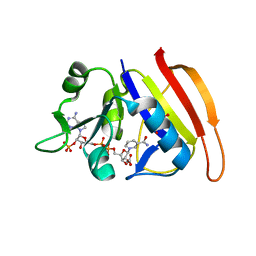 | |
2EW0
 
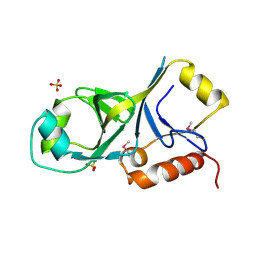 | | X-ray Crystal Structure of Protein Q6FF54 from Acinetobacter sp. ADP1. Northeast Structural Genomics Consortium Target AsR1. | | Descriptor: | SULFATE ION, hypothetical protein ACIAD0353 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Forouhar, F, Acton, T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel x-ray structure of hypothetical protein Q6FF54 at the resolution 1.4 A. Northeast Structural Genomics target AsR1.
TO BE PUBLISHED
|
|
3MAN
 
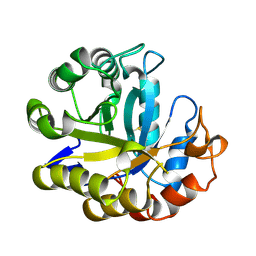 | | MANNOHEXAOSE COMPLEX OF THERMOMONOSPORA FUSCA BETA-MANNANASE | | Descriptor: | PROTEIN (BETA-MANNANASE), beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Hilge, M, Gloor, S.M, Piontek, K. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca - substrate specificity in glycosyl hydrolase family 5.
Structure, 6, 1998
|
|
3IG4
 
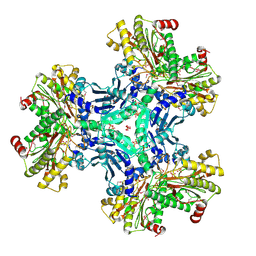 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
