8EMQ
 
 | | Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-28 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.66 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
8EN7
 
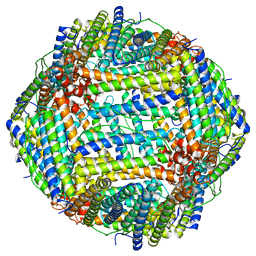 | | Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-28 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.68 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
5C1M
 
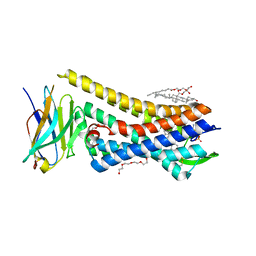 | | Crystal structure of active mu-opioid receptor bound to the agonist BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Huang, W.J, Manglik, A, Venkatakrishnan, A.J, Laeremans, T, Feinberg, E.N, Sanborn, A.L, Kato, H.E, Livingston, K.E, Thorsen, T.S, Kling, R, Granier, S, Gmeiner, P, Husbands, S.M, Traynor, J.R, Weis, W.I, Steyaert, J, Dror, R.O, Kobilka, B.K. | | Deposit date: | 2015-06-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into mu-opioid receptor activation.
Nature, 524, 2015
|
|
1TP8
 
 | | CRYSTAL STRUCTURE OF A GALACTOSE SPECIFIC LECTIN FROM ARTOCARPUS HIRSUTA IN COMPLEX WITH METHYL-a-D-GALACTOSE | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA CHAIN, methyl alpha-D-galactopyranoside | | Authors: | Rao, K.N, Suresh, C.G, Katre, U.V, Gaikwad, S.M, Khan, M.I. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two orthorhombic crystal structures of a galactose-specific lectin from Artocarpus hirsuta in complex with methyl-alpha-D-galactose.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8BZS
 
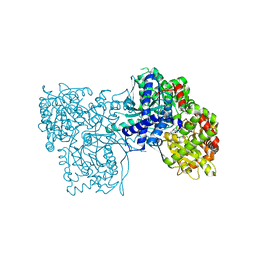 | | The crystal structure of glycogen phosphorylase in complex with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Koulas, S.M, Mathomes, R, Hayes, J.M, Leonidas, D.D. | | Deposit date: | 2022-12-15 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Multidisciplinary docking, kinetics and X-ray crystallography studies of baicalein acting as a glycogen phosphorylase inhibitor and determination of its' potential against glioblastoma in cellular models.
Chem.Biol.Interact., 382, 2023
|
|
1K88
 
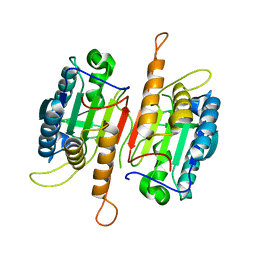 | | Crystal structure of procaspase-7 | | Descriptor: | procaspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
6U3V
 
 | |
3KTU
 
 | |
3EXZ
 
 | | Crystal structure of the MaoC-like dehydratase from Rhodospirillum rubrum. Northeast Structural Genomics Consortium target RrR103a. | | Descriptor: | MaoC-like dehydratase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-17 | | Release date: | 2008-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MaoC-like dehydratase from Rhodospirillum rubrum.
To be Published
|
|
3OGG
 
 | | Crystal structure of the receptor binding domain of botulinum neurotoxin D | | Descriptor: | Botulinum neurotoxin type D | | Authors: | Zhang, Y, Gao, X, Qin, L, Buchko, G.W, Robinson, H, Varnum, S.M. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural analysis of the receptor binding domain of botulinum neurotoxin serotype D.
Biochem.Biophys.Res.Commun., 401, 2010
|
|
6LNN
 
 | |
6LNU
 
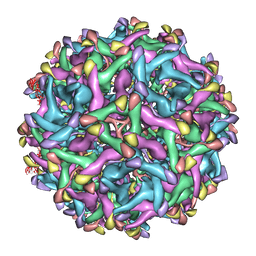 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
3FB2
 
 | | Crystal structure of the human brain alpha spectrin repeats 15 and 16. Northeast Structural Genomics Consortium target HR5563a. | | Descriptor: | Spectrin alpha chain, brain spectrin | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human brain alpha spectrin repeats 15 and 16.
To be Published
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
3KSF
 
 | |
1KEF
 
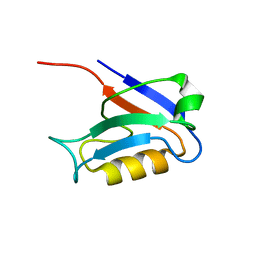 | | PDZ1 of SAP90 | | Descriptor: | synapse associated protein-90 | | Authors: | Piserchio, A, Pellegrini, M, Mehta, S, Blackman, S.M, Garcia, E.P, Marshall, J, Mierke, D.F. | | Deposit date: | 2001-11-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The PDZ1 domain of SAP90. Characterization of structure and binding.
J.Biol.Chem., 277, 2002
|
|
3KSI
 
 | |
5YN8
 
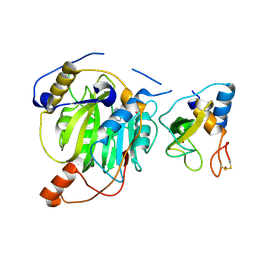 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNJ
 
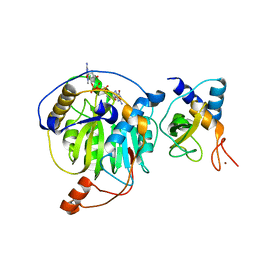 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNO
 
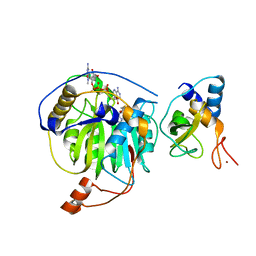 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
4XNN
 
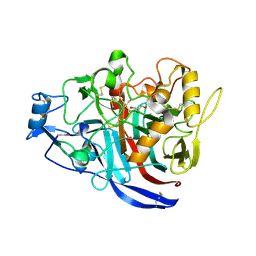 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Daphnia pulex | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL | | Authors: | Ebrahim, A, Hobdey, S.E, Podkaminer, K, Taylor II, L.E, Beckham, G.T, Decker, S.R, Himmel, M.E, Cragg, S.M, McGeehan, J.E. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a GH7 Family Cellobiohydrolase from Daphnia pulex
To Be Published
|
|
3KSG
 
 | |
4XDN
 
 | | Crystal structure of Scc4 in complex with Scc2n | | Descriptor: | MAU2 chromatid cohesion factor homolog, SULFATE ION, Sister chromatid cohesion protein 2 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural evidence for Scc4-dependent localization of cohesin loading.
Elife, 4, 2015
|
|
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
