2ZB8
 
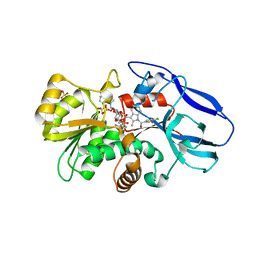 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and indomethacin | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZB3
 
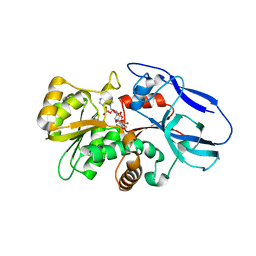 | | Crystal structure of mouse 15-ketoprostaglandin delta-13-reductase in complex with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
3CTN
 
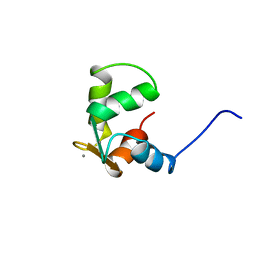 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
2YF3
 
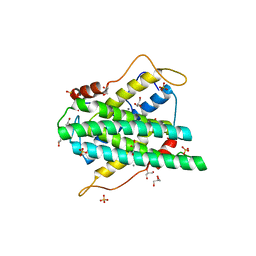 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, complex with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, ... | | Authors: | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
3DCD
 
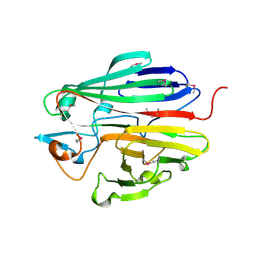 | | X-ray structure of the galactose mutarotase related enzyme Q5FKD7 from Lactobacillus acidophilus at the resolution 1.9A. Northeast Structural Genomics consortium target LaR33. | | Descriptor: | Galactose mutarotase related enzyme | | Authors: | Kuzin, A.P, Lew, S, Vorobiev, S.M, Seetharaman, J, Wang, H, Mao, L, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the galactose mutarotase related enzyme Q5FKD7 from Lactobacillus acidophilus at the resolution 1.9A. Northeast Structural Genomics consortium target LaR33. (CASP Target)
To be Published
|
|
5GNH
 
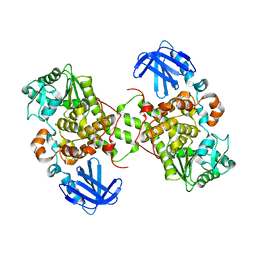 | | Myotubularin-related protein 2 | | Descriptor: | Myotubularin-related protein 2, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of MTMR2
To Be Published
|
|
5G22
 
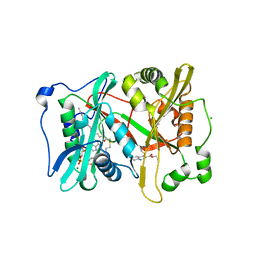 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 26) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
2YF9
 
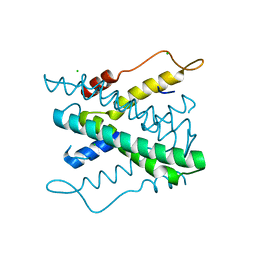 | | STRUCTURAL AND FUNCTIONAL INSIGHTS OF DR2231 PROTEIN, THE MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE FROM DEINOCOCCUS RADIODURANS, NATIVE FORM | | Descriptor: | CHLORIDE ION, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Authors: | Goncalves, A.M.D, De Sanctis, D, Mcsweeney, S.M. | | Deposit date: | 2011-04-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
1FT0
 
 | | CRYSTAL STRUCTURE OF TRUNCATED HUMAN RHOGDI K113A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3DAS
 
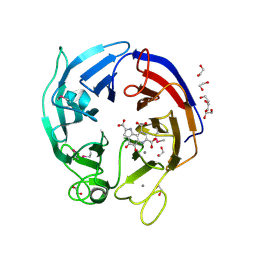 | | Structure of the PQQ-bound form of Aldose Sugar Dehydrogenase (Adh) from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Southall, S.M, Doel, J.J, Oubrie, A, Richardson, D.J. | | Deposit date: | 2008-05-30 | | Release date: | 2009-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Enzymatic Characterization of a Thermostable PQQ-dependent Soluble Aldose Sugar Dehydrogenase
To be Published
|
|
2YEZ
 
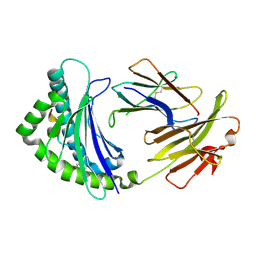 | | COMPLEX OF A B21 CHICKEN MHC CLASS I MOLECULE AND A 10MER CHICKEN PEPTIDE | | Descriptor: | 14-3-3 PROTEIN THETA, BETA-2-MICROGLOBULIN, MAJOR HISTOCOMPATIBILITY COMPLEX CLASS I GLYCOPROTEIN HAPLOTYPE B21 | | Authors: | Chappell, P, Roversi, P, Harrison, M.C, Mears, L.E, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Elife, 4, 2015
|
|
3DC7
 
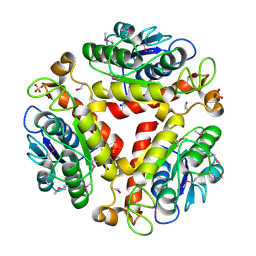 | | Crystal structure of the protein Q88SR8 from Lactobacillus plantarum. Northeast Structural Genomics consortium target LpR109. | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein lp_3323, SODIUM ION, ... | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Seetharaman, J, Zhao, L, Mao, L, Ciccosanti, C, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the protein Q88SR8 from Lactobacillus plantarum. Northeast Structural Genomics consortium target LpR109. (CASP Target)
To be Published
|
|
2XZ7
 
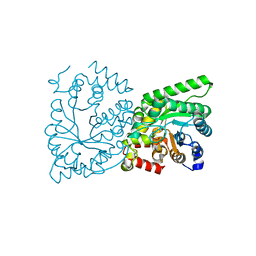 | | CRYSTAL STRUCTURE OF THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PHOSPHOENOLPYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA) | | Authors: | Waltersperger, S.M, Oberholzer, A.E, Schneider, P, Baumann, U, Erni, B. | | Deposit date: | 2010-11-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
4M32
 
 | | Crystal structure of gated-pore mutant D138N of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
5FMA
 
 | | human Notch 1, EGF 4-7 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Weisshuhn, P.C, Sheppard, D, Taylor, P, Whiteman, P, Lea, S.M, Handford, P.A, Redfield, C. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Non-Linear and Flexible Regions of the Human Notch1 Extracellular Domain Revealed by High-Resolution Structural Studies.
Structure, 24, 2016
|
|
1FSO
 
 | | CRYSTAL STRUCTURE OF TRUNCATED HUMAN RHOGDI QUADRUPLE MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2XX5
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (5E,10R)-N-BENZYL-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDRO-2-BENZAZACYCLOTETRADECINE-10-CARBOXAMIDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
1FI8
 
 | | RAT GRANZYME B [N66Q] COMPLEXED TO ECOTIN [81-84 IEPD] | | Descriptor: | ECOTIN, NATURAL KILLER CELL PROTEASE 1 | | Authors: | Waugh, S.M, Harris, J.L, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2000-08-03 | | Release date: | 2000-09-13 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the pro-apoptotic protease granzyme B reveals the molecular determinants of its specificity
Nat.Struct.Biol., 7, 2000
|
|
4MAC
 
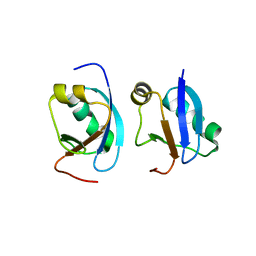 | | Crystal structure of CIDE-N domain of FSP27 | | Descriptor: | Cell death activator CIDE-3 | | Authors: | Park, H.H, Lee, S.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for homo-dimerization of the CIDE domain revealed by the crystal structure of the CIDE-N domain of FSP27
Biochem.Biophys.Res.Commun., 439, 2013
|
|
2XXS
 
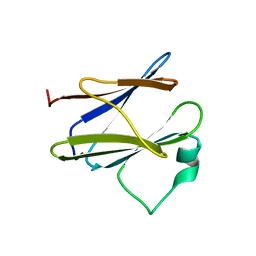 | | Solution structure of the N-terminal domain of the Shigella type III secretion protein MxiG | | Descriptor: | PROTEIN MXIG | | Authors: | McDowell, M.A, Johnson, S, Deane, J.E, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2010-11-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
J.Biol.Chem., 286, 2011
|
|
4LU3
 
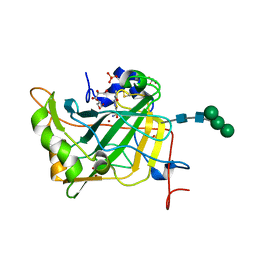 | | The crystal structure of the human carbonic anhydrase XIV | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 14, GLYCEROL, ... | | Authors: | Alterio, V, De Simone, G, Monti, S.M. | | Deposit date: | 2013-07-24 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural comparison between membrane-associated human carbonic anhydrases provides insights into drug design of selective inhibitors.
Biopolymers, 101, 2014
|
|
3D7M
 
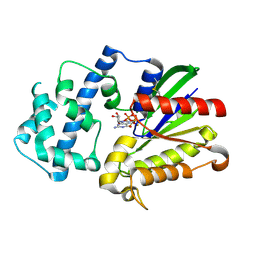 | | Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Funk, M.A, Preininger, A.M, Oldham, W.M, Meier, S.M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Helix dipole movement and conformational variability contribute to allosteric GDP release in Galphai subunits.
Biochemistry, 48, 2009
|
|
6UQC
 
 | | Mouse IgG2a Bispecific Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Tsai, J.C, Davis, J.H, West, S.M, Strop, P. | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design and characterization of mouse IgG1 and IgG2a bispecific antibodies for use in syngeneic models.
Mabs, 12, 2019
|
|
5G20
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 19). | | Descriptor: | 6-(BENZYLOXY)-4-(ETHYLSULFANYL)-3-[(MORPHOLIN-4-YL), DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
4KWJ
 
 | | Resting state of rat cysteine dioxygenase | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Souness, R.J, Wilbanks, S.M, Jameson, G.B, Jameson, G.N.L. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic implications of persulfenate and persulfide binding in the active site of cysteine dioxygenase.
Biochemistry, 52, 2013
|
|
