1O0V
 
 | | The crystal structure of IgE Fc reveals an asymmetrically bent conformation | | Descriptor: | GLYCEROL, Immunoglobulin heavy chain epsilon-1, SULFATE ION, ... | | Authors: | Wan, T, Beavil, R.L, Fabiane, S.M, Beavil, A.J, Sohi, M.K, Keown, M, Young, R.J, Henry, A.J, Owens, R.J, Gould, H.J, Sutton, B.J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of IgE Fc reveals an asymmetrically bent conformation
NAT.IMMUNOL., 3, 2002
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
1KEF
 
 | | PDZ1 of SAP90 | | Descriptor: | synapse associated protein-90 | | Authors: | Piserchio, A, Pellegrini, M, Mehta, S, Blackman, S.M, Garcia, E.P, Marshall, J, Mierke, D.F. | | Deposit date: | 2001-11-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The PDZ1 domain of SAP90. Characterization of structure and binding.
J.Biol.Chem., 277, 2002
|
|
1NVQ
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NR0
 
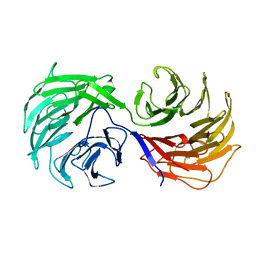 | | Two Seven-Bladed Beta-Propeller Domains Revealed By The Structure Of A C. elegans Homologue Of Yeast Actin Interacting Protein 1 (AIP1). | | Descriptor: | Actin interacting protein 1, MANGANESE (II) ION | | Authors: | Vorobiev, S.M, Mohri, K, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Functional Residues on Caenorhabditis elegans Actin-interacting Protein 1 (UNC-78) for Disassembly of Actin Depolymerizing Factor/Cofilin-bound Actin Filaments
J.Biol.Chem., 279, 2004
|
|
1NVX
 
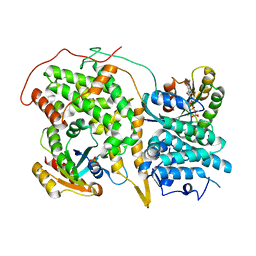 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
1NVW
 
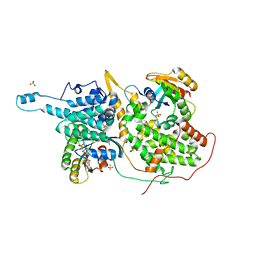 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
1NVR
 
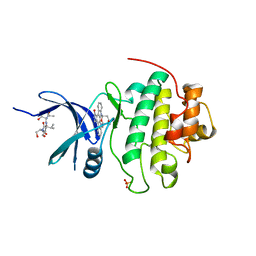 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NW9
 
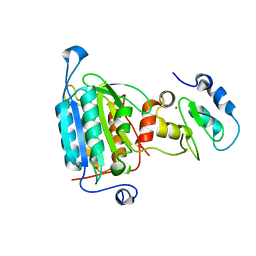 | | STRUCTURE OF CASPASE-9 IN AN INHIBITORY COMPLEX WITH XIAP-BIR3 | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION, caspase 9, ... | | Authors: | Shiozaki, E.N, Chai, J, Rigotti, D.J, Riedl, S.J, Li, P, Srinivasula, S.M, Alnemri, E.S, Fairman, R, Shi, Y. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of XIAP-Mediated Inhibition of Caspase-9
Mol.Cell, 11, 2003
|
|
1NLV
 
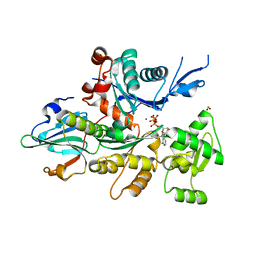 | | Crystal Structure Of Dictyostelium Discoideum Actin Complexed With Ca ATP And Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Vorobiev, S.M, Welti, S, Condeelis, J, Almo, S.C. | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Non-Vertebrate Actin: Implications For The ATP Hydrolytic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NVU
 
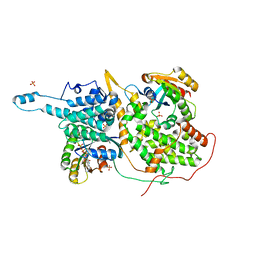 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
2V6L
 
 | | Molecular Model of a Type III Secretion System Needle | | Descriptor: | MXIH | | Authors: | Deane, J.E, Roversi, P, Cordes, F.S, Johnson, S, Kenjale, R, Daniell, S, Booy, F, Picking, W.L, Picking, W.D, Blocker, A.J, Lea, S.M. | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Molecular model of a type III secretion system needle: Implications for host-cell sensing.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
1DS9
 
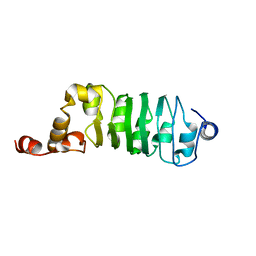 | | SOLUTION STRUCTURE OF CHLAMYDOMONAS OUTER ARM DYNEIN LIGHT CHAIN 1 | | Descriptor: | OUTER ARM DYNEIN | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2000-01-07 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a dynein motor domain associated light chain.
Nat.Struct.Biol., 7, 2000
|
|
2PBO
 
 | |
1BA1
 
 | |
1EQP
 
 | |
1AMW
 
 | |
2O7A
 
 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
3RU4
 
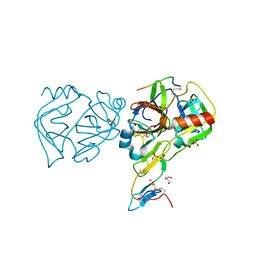 | | Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, Bowman-Birk type seed trypsin and chymotrypsin inhibitor, ... | | Authors: | Esteves, G.F, Santos, C.R, Ventura, M.M, Barbosa, J.A.R.G, Freitas, S.M. | | Deposit date: | 2011-05-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin
To be Published
|
|
2NQY
 
 | |
2OE9
 
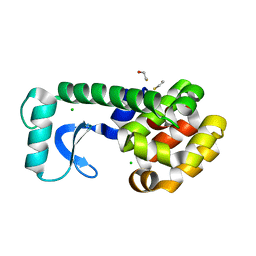 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
4LTM
 
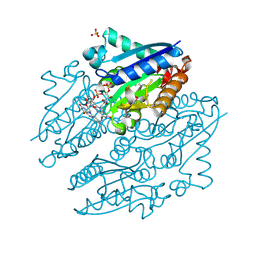 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
