5U0J
 
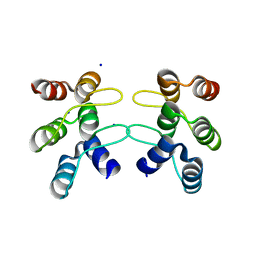 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - monoclinic crystal form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SODIUM ION | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5TSQ
 
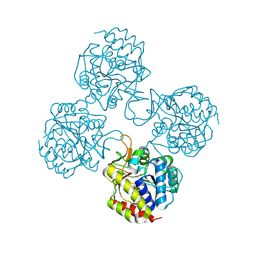 | | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose | | Descriptor: | CALCIUM ION, IUnH, beta-D-ribofuranose | | Authors: | Bachega, J.F.R, Timmers, L.F.S.M, Dalberto, P.F, Martinelli, L, Pinto, A.F.M, Basso, L.A, Norberto de Souza, O, Santos, D.S. | | Deposit date: | 2016-10-31 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose
To Be Published
|
|
5ZRP
 
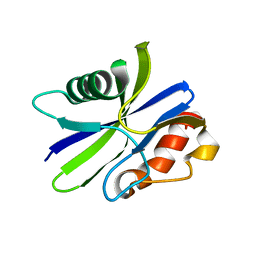 | | M. smegmatis antimutator protein MutT2 form 3 | | Descriptor: | Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRK
 
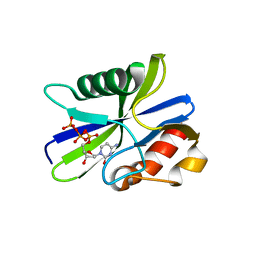 | | M. smegmatis antimutator protein MutT2 in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRI
 
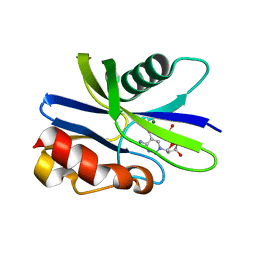 | | M. smegmatis antimutator protein MutT2 in complex with 5m-dCMP | | Descriptor: | MAGNESIUM ION, Putative mutator protein MutT2/NUDIX hydrolase, [(2R,3S,5R)-5-(4-azanyl-5-methyl-pyrimidin-1-ium-1-yl)-3-oxidanyl-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRC
 
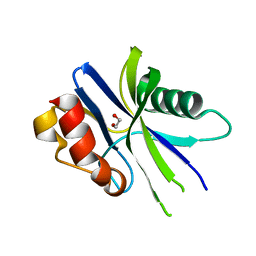 | | Structural insights into the catalysis mechanism of M. smegmatis antimutator protein MutT2 | | Descriptor: | 1,2-ETHANEDIOL, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRH
 
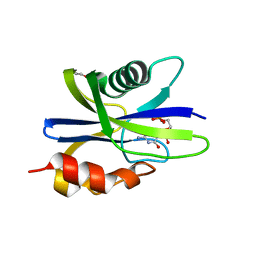 | | M. smegmatis antimutator protein MutT2 in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-MONOPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
8TID
 
 | | Combined linker domain of N-DRC and associated proteins Tetrahymena | | Descriptor: | AAA family ATPase CDC48 subfamily protein, CFAP20, Calmodulin 7-2, ... | | Authors: | Ghanaeian, A.G, Majhi, S.M, McCaffrey, C.M, Nami, B.N, Black, C.B, Yang, S.K, Legal, T.L, Papoulas, O.P, Janowska, M.J, Valente-Paterno, M.V, Marcotte, E.M, Wloga, D.W, Bui, K.H. | | Deposit date: | 2023-07-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
5ZRL
 
 | | M. smegmatis antimutator protein MutT2 in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-DIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
4ZP8
 
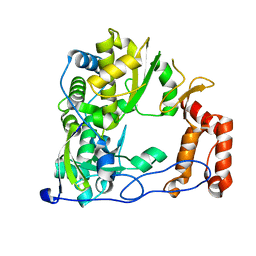 | | Coxsackievirus B3 Polymerase - F364L mutant | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4ZP9
 
 | | Coxsackievirus B3 Polymerase - F364I mutant | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4ZP6
 
 | | Coxsackievirus B3 Polymerase - F364A mutant | | Descriptor: | Genome polyprotein, SULFATE ION | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
6CPB
 
 | |
8F7M
 
 | |
4ZPA
 
 | | Coxsackievirus B3 Polymerase - F364Y mutant | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
2WQF
 
 | | Crystal Structure of the Nitroreductase CinD from Lactococcus lactis in Complex with FMN | | Descriptor: | COPPER INDUCED NITROREDUCTASE D, FLAVIN MONONUCLEOTIDE | | Authors: | Waltersperger, S.M, Oberholzer, A.E, Solioz, M, Baumann, U. | | Deposit date: | 2009-08-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Function of Cind (Ytjd) of Lactococcus Lactis, a Copper-Induced Nitroreductase Involved in Defense Against Oxidative Stress.
J.Bacteriol., 192, 2010
|
|
5DD0
 
 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB HEAVY LIGHT, oligo peptide | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
6BJM
 
 | | Human ABO(H) blood group glycosyltransferase GTB R188K mutant | | Descriptor: | ABO blood group (Transferase A, alpha 1-3-N-acetylgalactosaminyltransferase transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Gagnon, S.M.L, Legg, M.S.G, Evans, S.V. | | Deposit date: | 2017-11-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conserved residues Arg188 and Asp302 are critical for active site organization and catalysis in human ABO(H) blood group A and B glycosyltransferases.
Glycobiology, 28, 2018
|
|
5ZRO
 
 | | M. smegmatis antimutator protein MutT2 in complex with 5mdCTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
4ZP7
 
 | | Coxsackievirus B3 Polymerase - F364V mutant | | Descriptor: | Genome polyprotein | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4M53
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDPCP | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Arkhipova, V.I, Lazopulo, A.M, Lazopulo, S.M, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2013-08-07 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational transitions in the gamma subunit of the archaeal translation initiation factor 2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1HWH
 
 | |
7MJ9
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived mutant IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, Insulin-like growth factor-binding protein-like 1 altered peptide, MHC class I antigen | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ6
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ7
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
