2NYV
 
 | | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus | | Descriptor: | Phosphoglycolate phosphatase | | Authors: | Ciatto, C, Min, T, Gorman, J, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus
To be Published
|
|
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | Descriptor: | Hbl B protein | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
2NWI
 
 | | Crystal structure of protein AF1396 from Archaeoglobus fulgidus, Pfam DUF98 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Sridhar, V, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hypothetical protein O28875 from Archaeoglobus fulgidus
To be Published
|
|
2NX2
 
 | | Crystal structure of protein ypsA from Bacillus subtilis, Pfam DUF1273 | | Descriptor: | Hypothetical protein ypsA | | Authors: | Ramagopal, U.A, Alvarado, J, Dickey, M, Reyes, C, Toro, R, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein ypsA from Bacillus subtilis.
To be Published
|
|
4KVZ
 
 | |
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
2O3A
 
 | | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus | | Descriptor: | UPF0106 protein AF_0751 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Slocombe, A, Sridhar, V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus
To be Published
|
|
2OGF
 
 | | Crystal structure of protein MJ0408 from Methanococcus jannaschii, Pfam DUF372 | | Descriptor: | 8-OXOGUANINE, GLYCEROL, Hypothetical protein MJ0408, ... | | Authors: | Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of hypothetical protein MJ0408 from Methanococcus jannaschii
To be Published
|
|
2OG9
 
 | |
2OGK
 
 | |
2NS9
 
 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
4JR8
 
 | |
2OKT
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, ligand-free form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Toro, R, Malashkevich, V, Sauder, J.M, Ozyurt, S, Smith, D, Dickey, M, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2O16
 
 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
2NLY
 
 | | Crystal structure of protein BH1492 from Bacillus halodurans, Pfam DUF610 | | Descriptor: | Divergent polysaccharide deacetylase hypothetical protein, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein BH1492 from Bacillus halodurans C-125
To be Published
|
|
2NLZ
 
 | | Crystal structure of cephalosporin acylase from Bacillus halodurans | | Descriptor: | Cephalosporin acylase | | Authors: | Patskovsky, Y, Ramagopal, U, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cephalosporin acylase from Bacillus halodurans
To be Published
|
|
2NNB
 
 | | The Q403K mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Anderson, J.L.R, Miles, C.S, Mowat, C.G, Reid, G.A, Chapman, S.K. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Q403K mutant heme domain of flavocytochrome P450 BM3
To be published
|
|
4KW3
 
 | |
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
4K5S
 
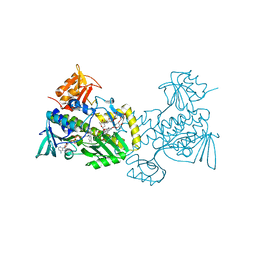 | | The crystal structure of premithramycin B in complex with MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase, premithramycin B | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
2OKV
 
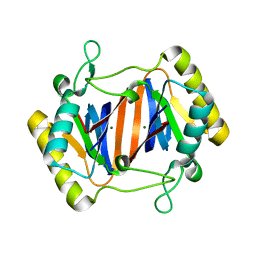 | | c-Myc DNA Unwinding Element Binding Protein | | Descriptor: | MAGNESIUM ION, Probable D-tyrosyl-tRNA(Tyr) deacylase 1 | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the c-myc DNA-unwinding Element-binding Protein DUE-B.
J.Biol.Chem., 282, 2007
|
|
2NUM
 
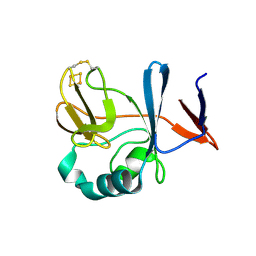 | | Soluble domain of Rieske Iron-Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-09 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NW7
 
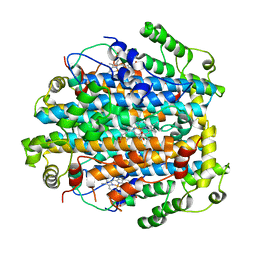 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferric heme. Northeast Structural Genomics Target XcR13 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Bruckmann, C, Thackray, S.J, Seetharaman, J, Tucker, T, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Chapman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NWU
 
 | |
2NQ5
 
 | | Crystal structure of methyltransferase from Streptococcus mutans | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of methyltransferase from Streptococcus mutans
To be Published
|
|
