2IMO
 
 | |
5KUI
 
 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with calcium. | | Descriptor: | Calcium uniporter protein, mitochondrial | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
2IMG
 
 | | Crystal structure of dual specificity protein phosphatase 23 from Homo sapiens in complex with ligand malate ion | | Descriptor: | D-MALATE, Dual specificity protein phosphatase 23 | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex.
J.Biol.Chem., 283, 2008
|
|
5KZ8
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 5,5-dimethyl-7-[(1~{S})-4-oxidanyl-1~{H}-inden-1-yl]-2-phenylazanyl-pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1YQP
 
 | | T268N mutant cytochrome domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
2IEC
 
 | | Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri | | Descriptor: | MAGNESIUM ION, Uncharacterized protein conserved in archaea | | Authors: | Bonanno, J.B, Ramagopal, U.A, Dickey, M, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri
TO BE PUBLISHED
|
|
5KNL
 
 | | Crystal structure of S. pombe ubiquitin E1 (Uba1) in complex with Ubc15 and ubiquitin | | Descriptor: | SULFATE ION, Ubiquitin, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Olsen, S.K, Lv, Z, Yuan, L, Williams, K. | | Deposit date: | 2016-06-28 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | S. pombe Uba1-Ubc15 Structure Reveals a Novel Regulatory Mechanism of Ubiquitin E2 Activity.
Mol. Cell, 65, 2017
|
|
2IM9
 
 | | Crystal structure of protein LPG0564 from Legionella pneumophila str. Philadelphia 1, Pfam DUF1460 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of hypothetical protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
TO BE PUBLISHED
|
|
2ISN
 
 | | Crystal structure of a phosphatase from a pathogenic strain Toxoplasma gondii | | Descriptor: | NYSGXRC-8828z, phosphatase, PRASEODYMIUM ION, ... | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IML
 
 | | Crystal structure of a hypothetical protein from Archaeoglobus fulgidus binding riboflavin 5'-phosphate | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a hypothetical protein from
Archaeoglobus fulgidus binding riboflavin 5'-phosphate
To be Published
|
|
2ICS
 
 | | Crystal structure of an adenine deaminase | | Descriptor: | ADENINE, Adenine Deaminase, ZINC ION | | Authors: | Sugadev, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine deaminase
TO BE PUBLISHED
|
|
2IM5
 
 | | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas Gingivalis | | Descriptor: | Nicotinate phosphoribosyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-03 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas gingivalis
To be Published
|
|
2IQI
 
 | | Crystal structure of protein XCC0632 from Xanthomonas campestris, Pfam DUF330 | | Descriptor: | Hypothetical protein XCC0632 | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Mckenzie, C, Pelletier, L, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein XCC0632 from Xanthomonas campestris pv. campestris
To be Published
|
|
7N1P
 
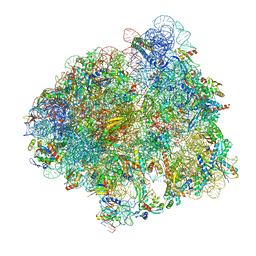 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
8FH2
 
 | |
8FGZ
 
 | |
2IJR
 
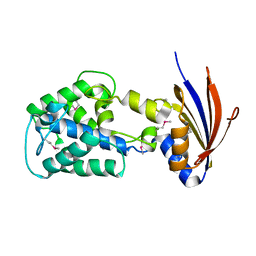 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
8FH1
 
 | |
8FH0
 
 | |
8FGY
 
 | |
5DZT
 
 | |
2IJQ
 
 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
4UBE
 
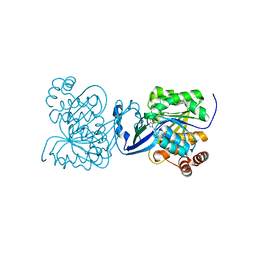 | | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C. | | Deposit date: | 2014-08-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE
TO BE PUBLISHED
|
|
4UNI
 
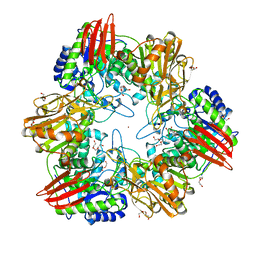 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
