3MOG
 
 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3QD4
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl{(3R,5R)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-5-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, SULFATE ION, tert-butyl {(3R,5R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-5-methylpiperidin-3-yl}carbamate | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3NAS
 
 | |
3NIV
 
 | |
3RO6
 
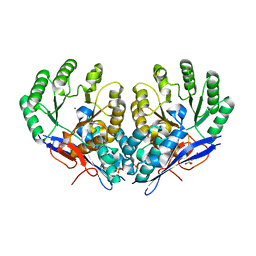 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg ion | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1C56
 
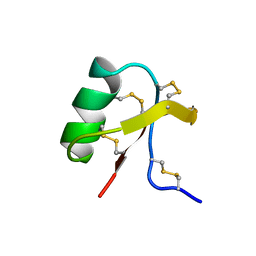 | |
3R03
 
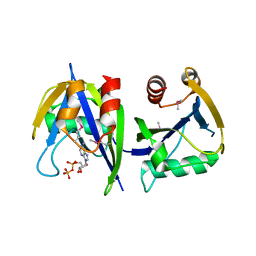 | |
3NSY
 
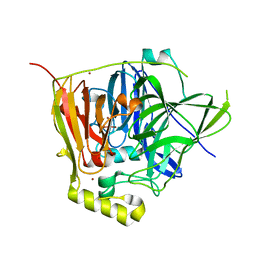 | | The multi-copper oxidase CueO with six Met to Ser mutations (M358S,M361S,M362S,M364S,M366S,M368S) | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
1C55
 
 | |
3MAB
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM LISTERIA MONOCYTOGENES, Triclinic FORM | | Descriptor: | uncharacterized protein | | Authors: | Madegowda, M, Chruszcz, M, Minor, W, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of an uncharacterized protein from listeria monocytogenes
To be Published
|
|
8THE
 
 | | Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme | | Descriptor: | (2~{R},4~{R},5~{S},6~{S})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{S})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Heme/hemoglobin uptake outer membrane receptor PhuR, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Knejski, P, Erramilli, S.K, Kossiakoff, A.A. | | Deposit date: | 2023-07-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Chaperone-assisted cryo-EM structure of P. aeruginosa PhuR reveals molecular basis for heme binding.
Structure, 32, 2024
|
|
3F4J
 
 | | Crystal structure of LeuT bound to glycine and sodium | | Descriptor: | GLYCINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
1CDW
 
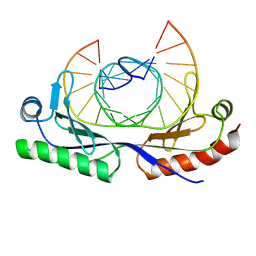 | | HUMAN TBP CORE DOMAIN COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*CP*TP*G)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)) | | Authors: | Nikolov, D.B, Chen, H, Halay, E.D, Hoffmann, A, Roeder, R.G, Burley, S.K. | | Deposit date: | 1996-04-11 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human TATA box-binding protein/TATA element complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3M9L
 
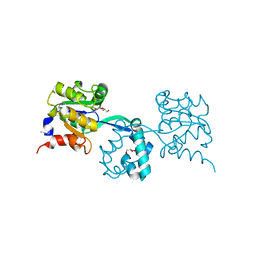 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 | | Descriptor: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Had Family Hydrolase from Pseudomonas Fluorescens Pf-5
To be Published
|
|
3M9U
 
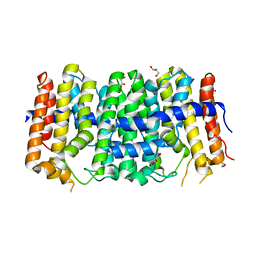 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Farnesyl-diphosphate synthase, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis Atcc 367
To be Published
|
|
3MJT
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
3MKC
 
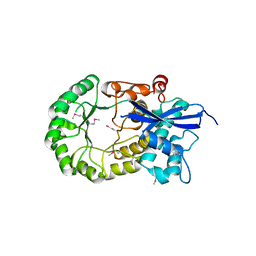 | |
3M2T
 
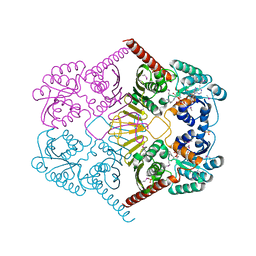 | |
3M3M
 
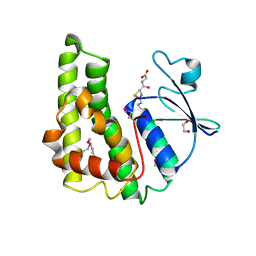 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
3MJC
 
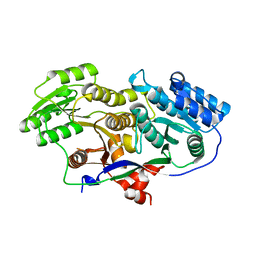 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, GLYCEROL | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
3MDN
 
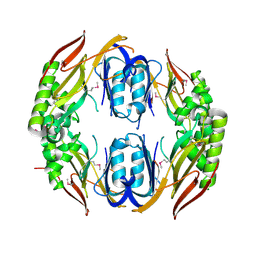 | |
3ME5
 
 | | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T | | Descriptor: | Cytosine-specific methyltransferase | | Authors: | Ramagopal, U.A, Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T
To be Published
|
|
3Q45
 
 | | Crystal structure of Dipeptide Epimerase from Cytophaga hutchinsonii complexed with Mg and dipeptide D-Ala-L-Val | | Descriptor: | D-ALANINE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family; possible chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2010-12-22 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NND
 
 | |
3R9E
 
 | |
