4XW1
 
 | |
3KLC
 
 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | ACETIC ACID, BROMIDE ION, Beta ureidopropionase (Beta-alanine synthase), ... | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-11-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
4W5Z
 
 | |
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
310D
 
 | | Crystal structure of 2'-O-Me(CGCGCG)2: an RNA duplex at 1.3 A resolution. Hydration pattern of 2'-O-methylated RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(OMC)P*(OMG)P*(OMC)P*(OMG)P*(OMC)P*(OMG))-3') | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Dauter, Z, Rypniewski, W.R. | | Deposit date: | 1996-05-25 | | Release date: | 1997-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of 2'-O-Me(CGCGCG)2, an RNA duplex at 1.30 A resolution. Hydration pattern of 2'-O-methylated RNA.
Nucleic Acids Res., 25, 1997
|
|
3IHI
 
 | | Crystal structure of mouse thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2009-07-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
1S3T
 
 | | BORATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | BORIC ACID, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Details of Urease Inhibition by Boric Acid: Insights into the Catalytic Mechanism.
J.Am.Chem.Soc., 126, 2004
|
|
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
5NOO
 
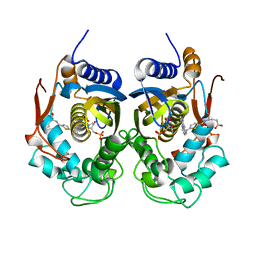 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP and Tomudex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, TOMUDEX, Thymidylate synthase | | Authors: | Wilk, P, Jarmula, A, Maj, P, Dowiercial, A, Banaszak, K, Rypniewski, W, Rode, W. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
3D7W
 
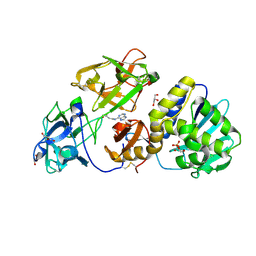 | | Mistletoe Lectin I in Complex with Zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, (2Z)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meyer, A, Rypniewski, W, Szymanski, M, Voelter, W, Barciszewski, J, Betzel, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of mistletoe lectin I from Viscum album in complex with the phytohormone zeatin
Biochim.Biophys.Acta, 1784, 2008
|
|
1Z24
 
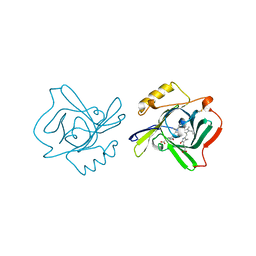 | | The molecular structure of insecticyanin from the tobacco hornworm Manduca sexta L. at 2.6 A resolution. | | Descriptor: | BILIVERDIN IX GAMMA CHROMOPHORE, Insecticyanin A form | | Authors: | Holden, H.M, Rypniewski, W.R, Law, J.H, Rayment, I. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of insecticyanin from the tobacco hornworm Manduca sexta L. at 2.6 A resolution.
Embo J., 6, 1987
|
|
4IRR
 
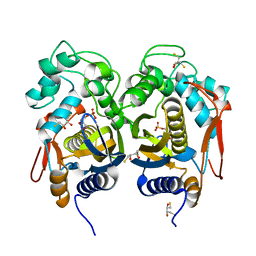 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP
To be Published
|
|
5BY6
 
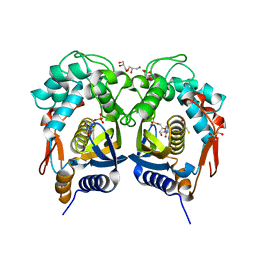 | | Crystal structure of Trichinella spiralis thymidylate synthase complexed with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Fraczyk, T, Wilk, P, Rode, W. | | Deposit date: | 2015-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
1I7J
 
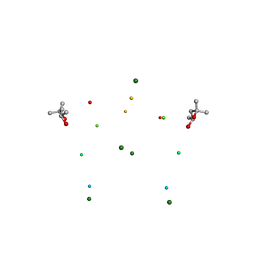 | | CRYSTAL STRUCTURE OF 2'-O-ME(CGCGCG)2: AN RNA DUPLEX AT 1.19 A RESOLUTION. 2-METHYL-2,4-PENTANEDIOL AND MAGNESIUM BINDING. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-R(*(OMC)P*(OMG)P*(OMC)P*(OMG)P*(OMC)P*(OMG))-3', MAGNESIUM ION | | Authors: | Adamiak, D.A, Rypniewski, W.R, Milecki, J, Adamiak, R.W. | | Deposit date: | 2001-03-09 | | Release date: | 2001-09-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | The 1.19 A X-ray structure of 2'-O-Me(CGCGCG)(2) duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove.
Nucleic Acids Res., 29, 2001
|
|
4EB4
 
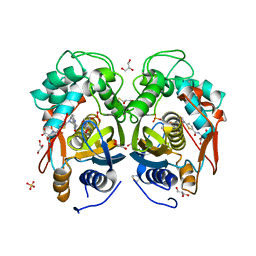 | | Crystal structure of mouse thymidylate synthase in ternary complex with dUMP and Tomudex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Wilk, P, Rode, W. | | Deposit date: | 2012-03-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of mouse thymidylate synthase in tertiary complex with dUMP and raltitrexed reveals N-terminus architecture and two different active site conformations.
Biomed Res Int, 2014, 2014
|
|
5MWI
 
 | |
4E5O
 
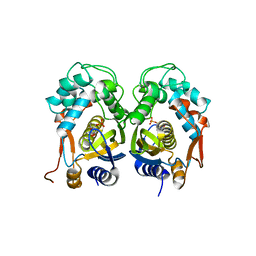 | | Crystal structure of mouse thymidylate synthase in complex with dUMP | | Descriptor: | 1,4-BUTANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
6QIS
 
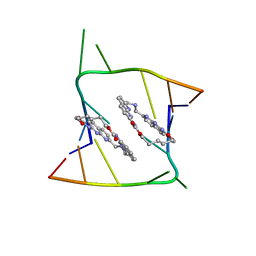 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model II) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3'), SULFATE ION | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
4MB4
 
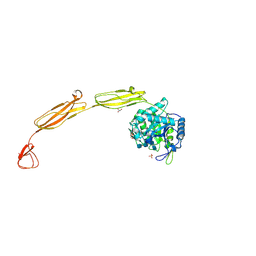 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
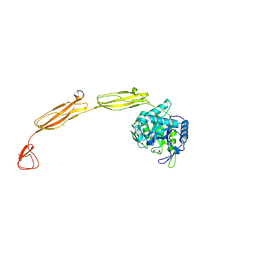 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2PUW
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, isomerase domain of glutamine-fructose-6-phosphate transaminase (isomerizing) | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
4MB5
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3R1C
 
 | | Crystal structure of GCGGCGGC duplex | | Descriptor: | RNA (5'-R(*GP*CP*GP*GP*CP*GP*GP*C)-3'), SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0531 Å) | | Cite: | Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome.
Nucleic Acids Res., 39, 2011
|
|
4HMD
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HME
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
