3EPT
 
 | |
3BMZ
 
 | | Violacein biosynthetic enzyme VioE | | Descriptor: | Putative uncharacterized protein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-12-13 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Violacein Biosynthetic Enzyme VioE Shares a Fold with Lipoprotein Transporter Proteins
J.Biol.Chem., 283, 2008
|
|
2R0G
 
 | | Chromopyrrolic acid-soaked RebC with bound 7-carboxy-K252c | | Descriptor: | 7-carboxy-5-hydroxy-12,13-dihydro-6H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, FLAVIN-ADENINE DINUCLEOTIDE, RebC | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0C
 
 | |
2R0P
 
 | | K252c-soaked RebC | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6XHM
 
 | | Covalent complex of SARS-CoV-2 main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHL
 
 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHO
 
 | | Covalent complex of SARS-CoV main protease with ethyl (4R)-4-({N-[(4-methoxy-1H-indol-2-yl)carbonyl]-L-leucyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHN
 
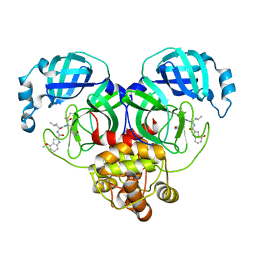 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Descriptor: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
4I23
 
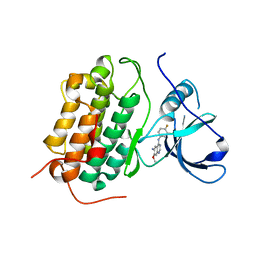 | | Crystal structure of the wild-type EGFR kinase domain in complex with dacomitinib (soaked) | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-methoxyquinazolin-6-yl}-4-(piperidin-1-yl)but-2-enamide, Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I21
 
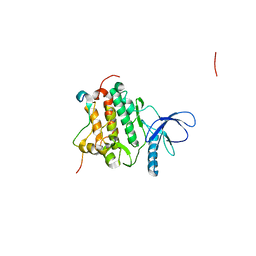 | | Crystal structure of L858R + T790M EGFR kinase domain in complex with MIG6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I20
 
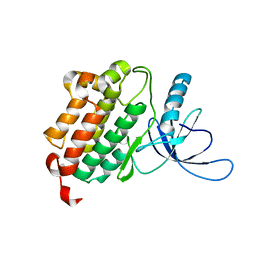 | | Crystal structure of monomeric (V948R) primary oncogenic mutant L858R EGFR kinase domain | | Descriptor: | Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I24
 
 | | Structure of T790M EGFR kinase domain co-crystallized with dacomitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-methoxyquinazolin-6-yl}-4-(piperidin-1-yl)but-2-enamide, Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I22
 
 | | Structure of the monomeric (V948R)gefitinib/erlotinib resistant double mutant (L858R+T790M) EGFR kinase domain co-crystallized with gefitinib | | Descriptor: | Epidermal growth factor receptor, Gefitinib, SULFATE ION | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I1Z
 
 | | Crystal structure of the monomeric (V948R) form of the gefitinib/erlotinib resistant EGFR kinase domain L858R+T790M | | Descriptor: | Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
7RF9
 
 | | O2-, PLP-dependent desaturase Plu4 intermediate-bound enzyme | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RGB
 
 | | O2-, PLP-dependent desaturase Plu4 product-bound enzyme | | Descriptor: | (2Z,4E)-5-carbamimidamido-2-iminopent-4-enoic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-14 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6C3C
 
 | | PLP-dependent L-arginine hydroxylase RohP quinonoid I complex | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6C3A
 
 | | O2-, PLP-dependent L-arginine hydroxylase RohP 4-hydroxy-2-ketoarginine complex | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
7KK6
 
 | |
7KK4
 
 | | Structure of the catalytic domain of PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK3
 
 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKP
 
 | |
7KKM
 
 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKO
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
