2Z5S
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
2Z5T
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
5ODN
 
 | |
5ODP
 
 | |
6HIT
 
 | | The crystal structure of haemoglobin from Atlantic cod | | Descriptor: | Hemoglobin alpha 2 chain, Hemoglobin beta 4 chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Helland, R, Bjorkeng, E.K, Rothweiler, U, Sydnes, M.O, Pampanin, D.M. | | Deposit date: | 2018-08-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of haemoglobin from Atlantic cod.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6KML
 
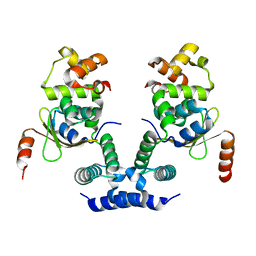 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KMQ
 
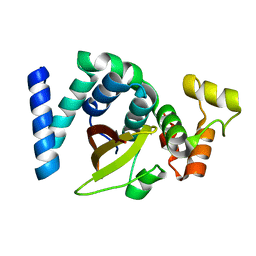 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
4D05
 
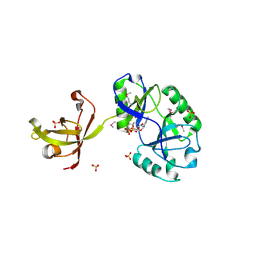 | | Structure and activity of a minimal-type ATP-dependent DNA ligase from a psychrotolerant bacterium | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT DNA LIGASE, MAGNESIUM ION, ... | | Authors: | Williamson, A, Rothweiler, U, Leiros, H.-K.S. | | Deposit date: | 2014-04-24 | | Release date: | 2014-11-12 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme-Adenylate Structure of a Bacterial ATP-Dependent DNA Ligase with a Minimized DNA-Binding Surface
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6F04
 
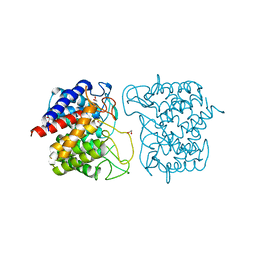 | | N-acetylglucosamine-2-epimerase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-acetylglucosamine-2-epimerase | | Authors: | Halsoer, M.J, Rothweiler, U. | | Deposit date: | 2017-11-17 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The crystal structure of the N-acetylglucosamine 2-epimerase from Nostoc sp. KVJ10 reveals the true dimer.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5N23
 
 | |
