2MLX
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA220-310 | | 分子名称: | Alkaline phosphatase, Trigger factor | | 著者 | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLY
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA1-150 | | 分子名称: | Alkaline phosphatase, Trigger factor | | 著者 | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLZ
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA365-471 | | 分子名称: | Alkaline phosphatase, Trigger factor | | 著者 | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2AOC
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | 分子名称: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
5KRW
 
 | | Recognition and targeting mechanisms by chaperones in flagella assembly and operation | | 分子名称: | Flagellar protein FliT,Flagellar hook-associated protein 2 fusion | | 著者 | Khanra, N.K, Rossi, P, Economou, A, Kalodimos, C.G. | | 登録日 | 2016-07-07 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recognition and targeting mechanisms by chaperones in flagellum assembly and operation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2AOD
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOE
 
 | | crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog CA-P2 | | 分子名称: | ACETIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AVV
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | 分子名称: | ACETIC ACID, CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | 著者 | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AOF
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P1-P6 | | 分子名称: | ACETIC ACID, CHLORIDE ION, PEPTIDE INHIBITOR, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOI
 
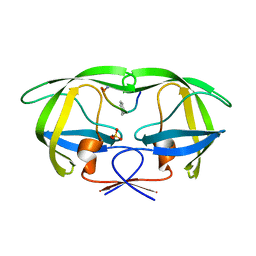 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | 分子名称: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AVM
 
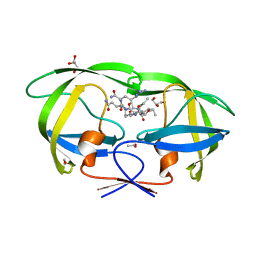 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | 分子名称: | ACETIC ACID, GLYCEROL, HIV-1 protease, ... | | 著者 | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AOH
 
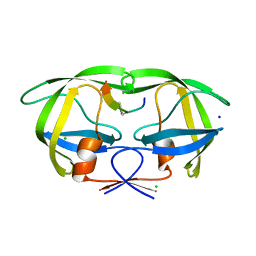 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | 分子名称: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
6PPT
 
 | |
6PQ2
 
 | |
6PQM
 
 | |
6PRQ
 
 | |
6PRJ
 
 | |
6PQE
 
 | |
6PRP
 
 | |
6PRI
 
 | |
6PSI
 
 | |
3MER
 
 | | Crystal Structure of the methyltransferase Slr1183 from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | 分子名称: | Slr1183 protein | | 著者 | Seetharaman, J, Chen, Y, Forouhar, F, Rossi, P, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-03-31 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target SgR145
To be Published
|
|
4O8W
 
 | | Crystal Structure of the GerD spore germination protein | | 分子名称: | Spore germination protein | | 著者 | Li, Y, Jin, K, Ghosh, S, Devarakonda, P, Carlson, K, Davis, A, Stewart, K, Cammett, E, Rossi, P.P, Setlow, B, Lu, M, Setlow, P, Hao, B. | | 登録日 | 2013-12-30 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.293 Å) | | 主引用文献 | Structural and Functional Analysis of the GerD Spore Germination Protein of Bacillus Species.
J.Mol.Biol., 426, 2014
|
|
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | 分子名称: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-11 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
