2SAM
 
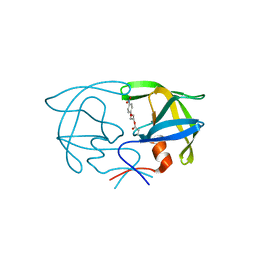 | | STRUCTURE OF THE PROTEASE FROM SIMIAN IMMUNODEFICIENCY VIRUS: COMPLEX WITH AN IRREVERSIBLE NON-PEPTIDE INHIBITOR | | Descriptor: | 3-(4-NITRO-PHENOXY)-PROPAN-1-OL, SIV PROTEASE | | Authors: | Rose, R.B, Rose, J.R, Salto, R, Craik, C.S, Stroud, R.M. | | Deposit date: | 1994-07-08 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protease from simian immunodeficiency virus: complex with an irreversible nonpeptide inhibitor.
Biochemistry, 32, 1993
|
|
1AZ5
 
 | |
1G39
 
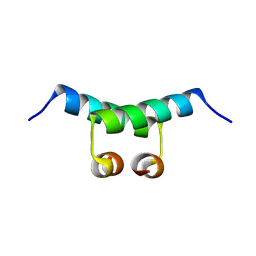 | | WILD-TYPE HNF-1ALPHA DIMERIZATION DOMAIN | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1G2Y
 
 | | HNF-1ALPHA DIMERIZATION DOMAIN, WITH SELENOMETHIONINE SUBSTITUED AT LEU 12 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1G2Z
 
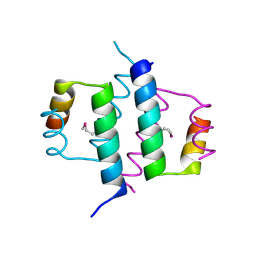 | | DIMERIZATION DOMAIN OF HNF-1ALPHA WITH A LEU 13 SELENOMETHIONINE SUBSTITUTION | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1RU0
 
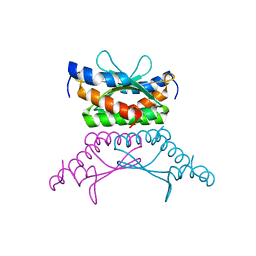 | | Crystal structure of DCoH2, a paralog of DCoH, the Dimerization Cofactor of HNF-1 | | Descriptor: | DcoH-like protein DCoHm | | Authors: | Rose, R.B, Pullen, K.E, Bayle, J.H, Crabtree, G.R, Alber, T. | | Deposit date: | 2003-12-10 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural basis for partially redundant enzymatic and transcriptional functions of DCoH and DCoH2
Biochemistry, 43, 2004
|
|
1F93
 
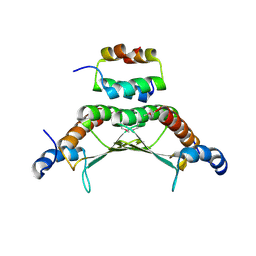 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE DIMERIZATION DOMAIN OF HNF-1 ALPHA AND THE COACTIVATOR DCOH | | Descriptor: | DIMERIZATION COFACTOR OF HEPATOCYTE NUCLEAR FACTOR 1-ALPHA, HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Bayle, J.H, Endrizzi, J.A, Cronk, J.D, Crabtree, G.R, Alber, T. | | Deposit date: | 2000-07-06 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1alpha.
Nat.Struct.Biol., 7, 2000
|
|
1YTG
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YTJ
 
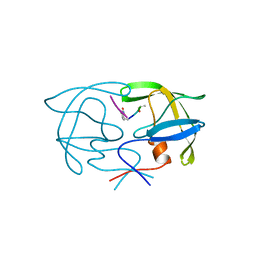 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | PEPTIDE PRODUCT, SIV PROTEASE | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YTH
 
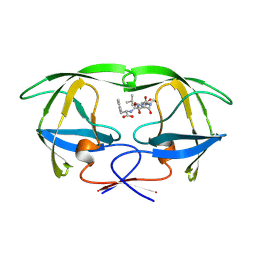 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YTI
 
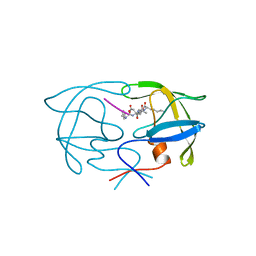 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | PEPTIDE PRODUCT, SIV PROTEASE | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
2QL2
 
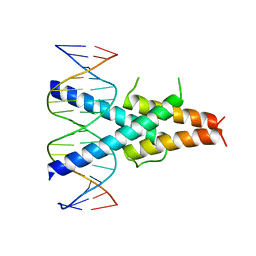 | | Crystal Structure of the basic-helix-loop-helix domains of the heterodimer E47/NeuroD1 bound to DNA | | Descriptor: | DNA (5'-D(*DAP*DGP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DTP*DGP*DGP*DCP*DCP*DTP*DA)-3'), DNA (5'-D(*DTP*DAP*DGP*DGP*DCP*DCP*DAP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DCP*DT)-3'), Neurogenic differentiation factor 1, ... | | Authors: | Rose, R.B, Longo, A, Guanga, G.P. | | Deposit date: | 2007-07-12 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition.
Biochemistry, 47, 2008
|
|
1W9G
 
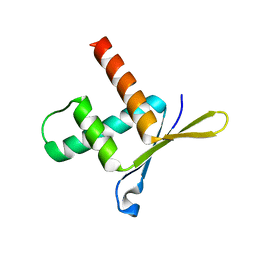 | | Structure of ERH (Enhencer of Rudimentary Gene) | | Descriptor: | ENHANCER OF RUDIMENTARY HOMOLOG | | Authors: | Wan, C, Tempel, W, Liu, Z, Wang, B.-C, Rose, R.B. | | Deposit date: | 2004-10-13 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Conserved Transcriptional Repressor Enhancer of Rudimentary Homolog
Biochemistry, 44, 2005
|
|
4WIL
 
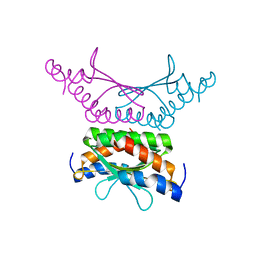 | | Crystal structure of DCoH2 S51T | | Descriptor: | Pterin-4-alpha-carbinolamine dehydratase 2 | | Authors: | Wang, D, Rose, R.B. | | Deposit date: | 2014-09-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Interactions with the Bifunctional Interface of the Transcriptional Coactivator DCoH1 Are Kinetically Regulated.
J.Biol.Chem., 290, 2015
|
|
3HXA
 
 | |
4EOT
 
 | | Crystal structure of the MafA homodimer bound to the consensus MARE | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*CP*CP*G)-3', 5'-D(*CP*CP*GP*GP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*GP*G)-3', SULFATE ION, ... | | Authors: | Lu, X, Guanga, G, Wan, C, Rose, R.B. | | Deposit date: | 2012-04-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | A Novel DNA Binding Mechanism for maf Basic Region-Leucine Zipper Factors Inferred from a MafA-DNA Complex Structure and Binding Specificities.
Biochemistry, 51, 2012
|
|
7KBL
 
 | | Biotin Carboxylase domain of Thermophilic 2-Oxoglutarate Carboxylase bound to Bicarbonate | | Descriptor: | 2-oxoglutarate carboxylase small subunit, BICARBONATE ION | | Authors: | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | Deposit date: | 2020-10-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
7KCT
 
 | | Crystal Structure of the Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase (OGC) Biotin Carboxylase (BC) Domain Dimer in Complex with Adenosine 5'-Diphosphate Magnesium Salt (MgADP), Adenosine 5'-Diphosphate (ADP, and Bicarbonate Anion (Hydrogen Carbonate/HCO3-) | | Descriptor: | 2-oxoglutarate carboxylase small subunit, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | Deposit date: | 2020-10-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
7KC7
 
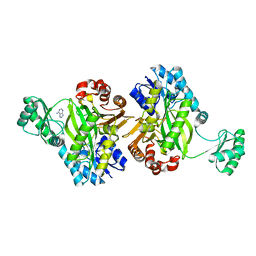 | | Biotin Carboxylase domain of Thermophilic 2-Oxoglutarate Carboxylase bound to ADP without Magnesium with disordered phosphate tail | | Descriptor: | 2-oxoglutarate carboxylase small subunit, ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | Deposit date: | 2020-10-05 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
7LHY
 
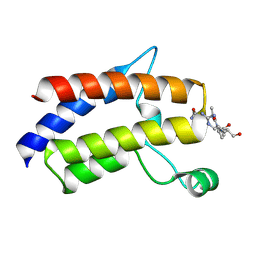 | |
2H1K
 
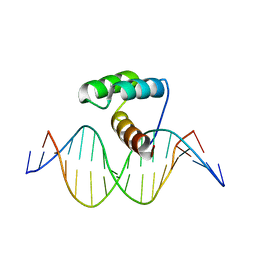 | |
1KGB
 
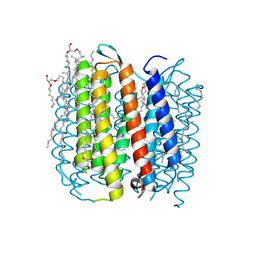 | | structure of ground-state bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1KG8
 
 | | X-ray structure of an early-M intermediate of bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1KG9
 
 | | Structure of a "mock-trapped" early-M intermediate of bacteriorhosopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
