2BN2
 
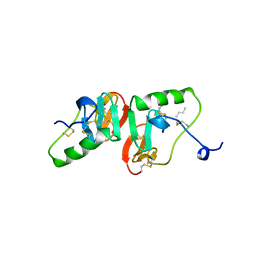 | |
1JK4
 
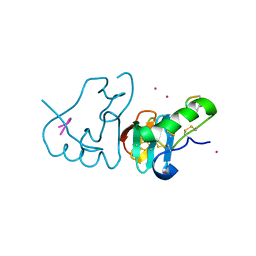 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
1JK6
 
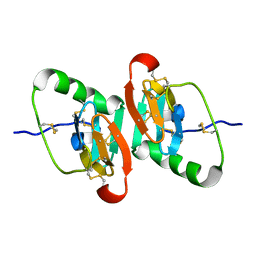 | | UNCOMPLEXED DES 1-6 BOVINE NEUROPHYSIN | | Descriptor: | NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2003
|
|
1OQC
 
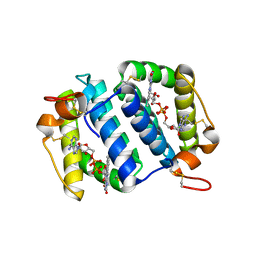 | |
1NPO
 
 | |
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4MN0
 
 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | Descriptor: | Berovin, CALCIUM ION, MAGNESIUM ION | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6B1B
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI MUTANT XS6 (APO Enzyme) | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, trimethylamine oxide | | Authors: | Zhou, D, Kandavelu, P, Wang, B.C, Yan, Y, Rose, J.P. | | Deposit date: | 2017-09-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
4TNO
 
 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4RNP
 
 | | BACTERIOPHAGE T7 RNA POLYMERASE, HIGH SALT CRYSTAL FORM, LOW TEMPERATURE DATA, ALPHA-CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Liu, Z.J, Sousa, R, Rose, J.P, Wang, B.C. | | Deposit date: | 1997-09-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution.
Nature, 364, 1993
|
|
1HRK
 
 | | CRYSTAL STRUCTURE OF HUMAN FERROCHELATASE | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, FERROCHELATASE | | Authors: | Wu, C.K, Dailey, H.A, Rose, J.P, Burden, A, Sellers, V.M, Wang, B.-C. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1PRY
 
 | | Structure Determination of Fibrillarin Homologue From Hyperthermophilic Archaeon Pyrococcus furiosus (Pfu-65527) | | Descriptor: | Fibrillarin-like pre-rRNA processing protein | | Authors: | Deng, L, Starostina, N.G, Liu, Z.-J, Rose, J.P, Terns, R.M, Terns, M.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-06-20 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus
Biochem.Biophys.Res.Commun., 315, 2004
|
|
1ZD0
 
 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
1I4W
 
 | | THE CRYSTAL STRUCTURE OF THE TRANSCRIPTION FACTOR SC-MTTFB OFFERS INTRIGUING INSIGHTS INTO MITOCHONDRIAL TRANSCRIPTION | | Descriptor: | MITOCHONDRIAL REPLICATION PROTEIN MTF1, XENON | | Authors: | Schubot, F.D, Chen, C.-J, Rose, J.P, Dailey, T.A, Dailey, H.A, Wang, B.-C. | | Deposit date: | 2001-02-23 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription.
Protein Sci., 10, 2001
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
3R7C
 
 | | The structure of a hexahestidine-tagged form of augmenter of liver regeneration reveals a novel Cd(2)Cl(4)O(6) cluster that aids in crystal packing | | Descriptor: | CADMIUM ION, CHLORIDE ION, FAD-linked sulfhydryl oxidase ALR, ... | | Authors: | Florence, Q.J, Wu, C.-K, Swindell II, J.T, Wang, B.C, Rose, J.P. | | Deposit date: | 2011-03-22 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a hexahestidine-tagged form of augmenter of liver regeneration reveals a novel Cd(2)Cl(4)O(6) cluster that aids in crystal packing
To be Published
|
|
1JF2
 
 | | Crystal Structure of W92F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, obelin | | Authors: | Liu, Z.-J, Vysotski, E.S, Deng, L, Markova, S.V, Lee, J, Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Violet bioluminescence and fast kinetics from W92F obelin: structure-based proposals for the bioluminescence triggering and the identification of the emitting species.
Biochemistry, 42, 2003
|
|
8SV7
 
 | |
2PNJ
 
 | | Crystal structure of human ferrochelatase mutant with Phe 337 replaced by Ala | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A.E, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis
Biochemistry, 46, 2007
|
|
2PO7
 
 | | Crystal structure of human ferrochelatase mutant with His 341 replaced by Cys | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis.
Biochemistry, 46, 2007
|
|
1AGS
 
 | |
1HQL
 
 | | The xenograft antigen in complex with the B4 isolectin of Griffonia simplicifolia lectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Tempel, W, Lipscomb, L.A, Rose, J.P, Woods, R.J. | | Deposit date: | 2000-12-18 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The xenograft antigen bound to Griffonia simplicifolia lectin 1-B(4). X-ray crystal structure of the complex and molecular dynamics characterization of the binding site.
J.Biol.Chem., 277, 2002
|
|
2A4K
 
 | | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137 | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase, UNKNOWN ATOM OR ION | | Authors: | Zhou, W, Ebihara, A, Tempel, W, Yokoyama, S, Chen, L, Kuramitsu, S, Nguyen, J, Chang, S.-H, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137
To be Published
|
|
3OV8
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus, High resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, Protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-09-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
