4RMW
 
 | |
5CSG
 
 | |
4RMU
 
 | |
4RMV
 
 | |
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
1A2S
 
 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGA MONORAPHIDIUM BRAUNII, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Banci, L, Bertini, I, De La Rosa, M.A, Koulougliotis, D, Navarro, J.A, Walter, O. | | Deposit date: | 1998-01-10 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized cytochrome c6 from the green alga Monoraphidium braunii.
Biochemistry, 37, 1998
|
|
4RMT
 
 | | Crystal structure of the D98N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | de Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | Decoding the Structural Bases of D76N 2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis.
Plos One, 10, 2015
|
|
4RMR
 
 | |
4RMQ
 
 | |
4RMS
 
 | |
1PCS
 
 | | THE 2.15 A CRYSTAL STRUCTURE OF A TRIPLE MUTANT PLASTOCYANIN FROM THE CYANOBACTERIUM SYNECHOCYSTIS SP. PCC 6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Romero, A, De La Cerda, B, Varela, P.F, Navarro, J.A, Hervas, M, De La Rosa, M.A. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The 2.15 A crystal structure of a triple mutant plastocyanin from the cyanobacterium Synechocystis sp. PCC 6803.
J.Mol.Biol., 275, 1998
|
|
4C7U
 
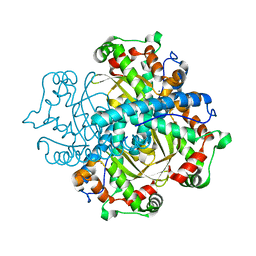 | | Crystal structure of manganese superoxide dismutase from Arabidopsis thaliana | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] 1, MITOCHONDRIAL | | Authors: | Marques, A, Santos, S.P, Rosa, M, Carrondo, M.A, Abreu, I.A, Romao, C.V, Frazao, C. | | Deposit date: | 2013-09-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Arabidopsis Thaliana Manganese Superoxide Dismutase at 1.95 A Resolution
To be Published
|
|
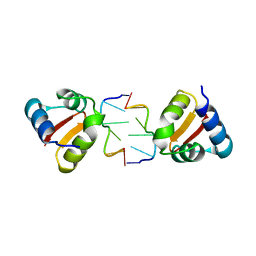
4KA4
 
 | |
4LB5
 
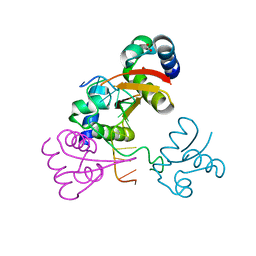 | |
4LB6
 
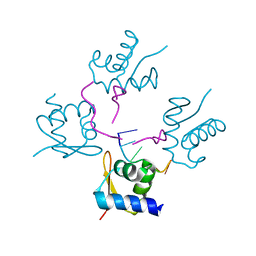 | |
4OJH
 
 | |
6H1F
 
 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
5KK3
 
 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
2F9R
 
 | | Crystal structure of the inactive state of the Smase I, a sphingomyelinase D from Loxosceles laeta venom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Sphingomyelinase D 1 | | Authors: | Murakami, M.T, Gabdoulkhakov, A, Fernandes-Pedrosa, M.F, Betzel, C, Tambourgi, D.V, Arni, R.K. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases D.
J.Biol.Chem., 280, 2005
|
|
3BHS
 
 | | Nitrosomonas europaea Rh50 and mechanism of conduction by Rhesus protein family of channels | | Descriptor: | Ammonium transporter family protein Rh50 | | Authors: | Gruswitz, F, Ho, C.-M, del Rosario, M.C, Westhoff, C.M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Nitrosomonas europaea Rh50 and mechanism of conduction by Rhesus protein family of channels.
To be Published
|
|
6VL2
 
 | | Stigmurin | | Descriptor: | Stigmurin | | Authors: | Rodrigues, S.C.S, Resende, J.M, Araujo, R.M, Pedrosa, M.F.F. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR three-dimensional structure of the cationic peptide Stigmurin from Tityus stigmurus scorpion venom: In vitro antioxidant and in vivo antibacterial and healing activity.
Peptides, 137, 2021
|
|
7KDQ
 
 | |
