7TOQ
 
 | | Mammalian 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein poly-PR | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
6TZA
 
 | |
6TZ5
 
 | |
7SSO
 
 | |
7SSW
 
 | |
7SSD
 
 | |
7SSL
 
 | |
8VS9
 
 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site and deacyl-tRNA in the E site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
8VSA
 
 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
4NS2
 
 | |
5CUH
 
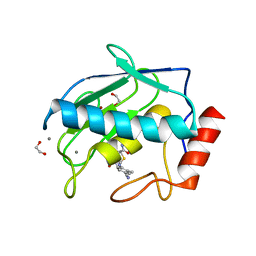 | | Crystal structure MMP-9 complexes with a constrained hydroxamate based inhibitor LT4 | | Descriptor: | (4S)-3-{[4-(4-cyano-2-methylphenyl)piperazin-1-yl]sulfonyl}-N-hydroxy-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tepshi, L, Vera, L, Nuti, E, Rosalia, L, Rossello, A, Stura, E.A. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a new selective inhibitor of A Disintegrin And Metalloprotease 10 (ADAM-10) able to reduce the shedding of NKG2D ligands in Hodgkin's lymphoma cell models.
Eur.J.Med.Chem., 111, 2016
|
|
4NVQ
 
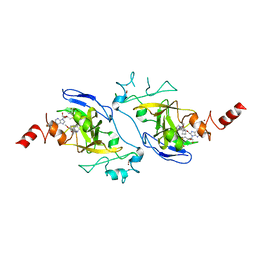 | | Human G9a in Complex with Inhibitor A-366 | | Descriptor: | 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Sweis, R.F, Pliushchev, M, Brown, P.J, Guo, J, Li, F, Maag, D, Petros, A.M, Soni, N.B, Tse, C, Vedadi, M, Michaelides, M.R, Chiang, G.G, Pappano, W.N. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and development of potent and selective inhibitors of histone methyltransferase g9a.
ACS Med Chem Lett, 5, 2014
|
|
6SME
 
 | |
9BF3
 
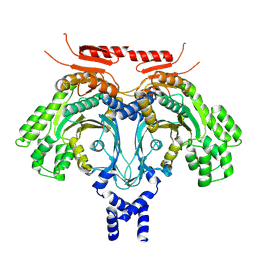 | | Cryo-EM Structure of GCN2 HRSL Domain | | Descriptor: | non-specific serine/threonine protein kinase | | Authors: | Solorio-Kirpichyan, K.M, Golovenko, D, Xiao, F, Yan, N, Korostelev, A.A, Korennykh, A.V. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM Structure of GCN2 HisRS-Like Domain
To Be Published
|
|
8TNV
 
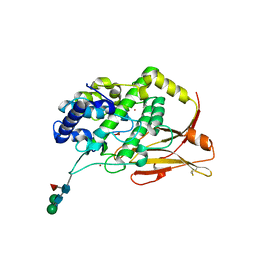 | | Hemocyanin Functional Unit CCHB-g of Concholepas concholepas | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Munoz, S, Vallejos-Baccelliere, G, Manubens, A, Salazar, M, Nascimento, A.F.Z, Ambrosio, A.L.B, Becker, M.I, Guixe, V, Castro-Fernandez, V. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a functional unit from an immunogenic mollusk hemocyanin.
Structure, 32, 2024
|
|
5NB4
 
 | | Atomic resolution structure of C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, HYDROGENPHOSPHATE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
9CL4
 
 | | Particulate methane monooxygenase in crosslinked, washed native membranes | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexadecanoyloxy)propyl (9Z)-heptadec-9-enoate, COPPER (II) ION, Particulate methane monooxygenase alpha subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9CL2
 
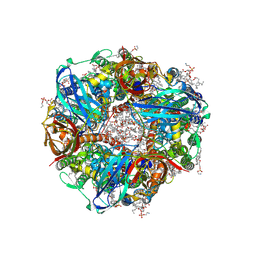 | | Particulate methane monooxygenase in washed native membranes | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexadecanoyloxy)propyl (9Z)-heptadec-9-enoate, COPPER (II) ION, Particulate methane monooxygenase alpha subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9CL3
 
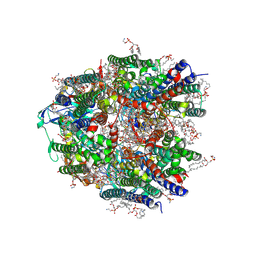 | | Particulate methane monooxygenase in unwashed native membranes | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexadecanoyloxy)propyl (9Z)-heptadec-9-enoate, COPPER (II) ION, Particulate methane monooxygenase alpha subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9C50
 
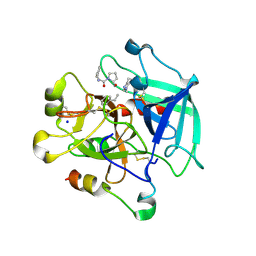 | | Replacement of a single residue changes the primary specificity of thrombin | | Descriptor: | FPF, SODIUM ION, Thrombin A-chain, ... | | Authors: | Dei Rossi, A, Deavila, S, Mohammed, B.M, Korolev, S, Di Cera, E. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Replacement of a single residue changes the primary specificity of thrombin.
J.Thromb.Haemost., 23, 2025
|
|
7Q4I
 
 | | Crystal structure of DmC1GalT1 in complex with UDP-Mn2+ and the APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1, ... | | Authors: | Gonzalez-Ramirez, A.M, Coelho, H, Companon, I, Grosso, A.S, Yang, Z, Narimatsu, Y, Clausen, H, Marcelo, F, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2021-10-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the synthesis of the core 1 structure by C1GalT1.
Nat Commun, 13, 2022
|
|
2O2F
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O21
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
7N0J
 
 | | Structure of YebY from E. coli K12 | | Descriptor: | YebY | | Authors: | Hadley, R.C, Rosenzweig, A.C. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The copper-linked Escherichia coli AZY operon: Structure, metal binding, and a possible physiological role in copper delivery.
J.Biol.Chem., 298, 2022
|
|
2O1Y
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
