5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPV
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPW
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPS
 
 | | Structure of RelA bound to ribosome in absence of A/R tRNA (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6AWB
 
 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWD
 
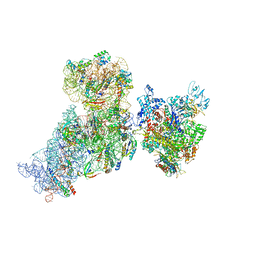 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
5U0K
 
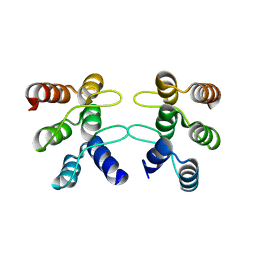 | | C-terminal ankyrin repeats from human liver-type glutaminase (GAB/LGA) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Ferreira, I.M, Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5U0J
 
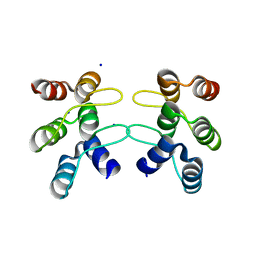 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - monoclinic crystal form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SODIUM ION | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
7ADO
 
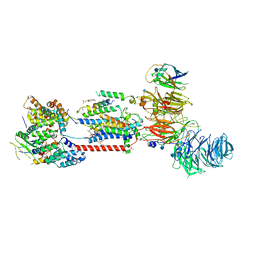 | | Cryo-EM structure of human ER membrane protein complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
3ZIZ
 
 | | Crystal structure of Podospora anserina GH5 beta-(1,4)-mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 ENDO-BETA-1,4-MANNANASE, GLYCEROL | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
5TWJ
 
 | | Crystal Structure of RlmH in Complex with S-Adenosylmethionine | | Descriptor: | Ribosomal RNA large subunit methyltransferase H, S-ADENOSYLMETHIONINE | | Authors: | Koh, C.S, Madireddy, R, Beane, T.J, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Small methyltransferase RlmH assembles a composite active site to methylate a ribosomal pseudouridine.
Sci Rep, 7, 2017
|
|
6BU8
 
 | | 70S ribosome with S1 domains 1 and 2 (Class 1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2017-12-08 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM.
Methods, 137, 2018
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
6BOK
 
 | |
6B4V
 
 | |
5TEG
 
 | |
5T7L
 
 | | Pt(II)-mediated copper-dependent interactions between ATOX1 and MNK1 | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1, ... | | Authors: | Caliandro, R, Mirabelli, V, Caliandro, R, Rosato, A, Lasorsa, A, Galliani, A, Arnesano, F, Natile, G. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic and Structural Basis for Inhibition of Copper Trafficking by Platinum Anticancer Drugs.
J.Am.Chem.Soc., 141, 2019
|
|
1YJU
 
 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJT
 
 | | Solution structure of the Cu(I) form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJR
 
 | | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJV
 
 | | Solution structure of the Cu(I) form of the sixth soluble domain of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
7ADP
 
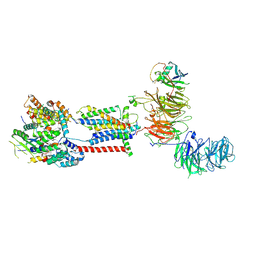 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
6DNC
 
 | | E.coli RF1 bound to E.coli 70S ribosome in response to UAU sense A-site codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Korostelev, A.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of premature translation termination on a sense codon.
J. Biol. Chem., 293, 2018
|
|
6BOH
 
 | |
3ZPS
 
 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[(4R)-5-[[(2S)-3,3-dimethyl-1-oxidanylidene-1-(prop-2-enylamino)butan-2-yl]amino]-4-oxidanyl-5-oxidanylidene-4-(phenylmethyl)pentyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors.
J.Med.Chem., 56, 2013
|
|
