3DSL
 
 | | The Three-dimensional Structure of Bothropasin, the Main Hemorrhagic Factor from Bothrops jararaca venom. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FUROYL-LEUCINE, ... | | Authors: | Muniz, J.R.C, Ambrosio, A, Selistre-de-Araujo, H.S, Oliva, G, Garratt, R.C, Souza, D.H.F. | | Deposit date: | 2008-07-13 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: Insights for a new classification of snake venom metalloprotease subgroups.
Toxicon, 52, 2008
|
|
4N83
 
 | |
4KRX
 
 | | Structure of Aes from E. coli | | Descriptor: | Acetyl esterase, TETRAETHYLENE GLYCOL | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
1YUB
 
 | | SOLUTION STRUCTURE OF AN RRNA METHYLTRANSFERASE (ERMAM) THAT CONFERS MACROLIDE-LINCOSAMIDE-STREPTOGRAMIN ANTIBIOTIC RESISTANCE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RRNA METHYLTRANSFERASE | | Authors: | Yu, L, Petros, A.M, Schnuchel, A, Zhong, P, Severin, J.M, Walter, K, Holzman, T.F, Fesik, S.W. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance.
Nat.Struct.Biol., 4, 1997
|
|
1IC0
 
 | | RED COPPER PROTEIN NITROSOCYANIN FROM NITROSOMONAS EUROPAEA | | Descriptor: | COPPER (II) ION, Nitrosocyanin | | Authors: | Lieberman, R.L, Arciero, D.M, Hooper, A.B, Rosenzweig, A.C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a novel red copper protein from Nitrosomonas europaea.
Biochemistry, 40, 2001
|
|
3TJK
 
 | | Crystal Structure of human peroxiredoxin IV C245A mutant in reduced form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJG
 
 | | Crystal Structure of human peroxiredoxin IV C51A mutant in oxidized form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3FEY
 
 | |
1JK9
 
 | | Heterodimer between H48F-ySOD1 and yCCS | | Descriptor: | SULFATE ION, ZINC ION, copper chaperone for superoxide dismutase, ... | | Authors: | Lamb, A.L, Torres, A.S, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Heterodimeric structure of superoxide dismutase in complex with its metallochaperone.
Nat.Struct.Biol., 8, 2001
|
|
3EWK
 
 | |
3J6Y
 
 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 2 degree rotation (Class I) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1YEW
 
 | |
3FBV
 
 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
3FDC
 
 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
4M1H
 
 | | X-ray crystal structure of Chlamydia trachomatis apo NrdB | | Descriptor: | Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Basis for Assembly of the Mn(IV)/Fe(III) Cofactor in the Class Ic Ribonucleotide Reductase from Chlamydia trachomatis.
Biochemistry, 52, 2013
|
|
3J6X
 
 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3FEX
 
 | |
4HMA
 
 | | Crystal structure of an MMP twin carboxylate based inhibitor LC20 in complex with the MMP-9 catalytic domain | | Descriptor: | CALCIUM ION, D-MALATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Antoni, C, Vera, L, Nuti, E, Carafa, L, Cassar-Lajeunesse, E, Dive, V, Rossello, A. | | Deposit date: | 2012-10-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4KRY
 
 | | Structure of Aes from E. coli in covalent complex with PMS | | Descriptor: | Acetyl esterase, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
3TJB
 
 | | Crystal structure of wild-type human peroxiredoxin IV | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
1W5W
 
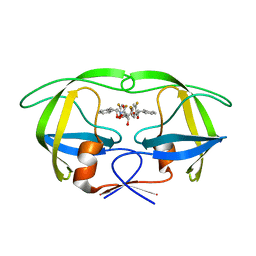 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,4-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1R,2S)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
3TJJ
 
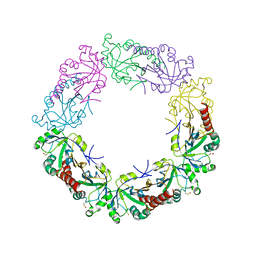 | | Crystal structure of human peroxiredoxin IV C245A mutant in sulfenylated form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJF
 
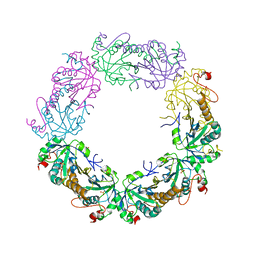 | | Crystal Structure of human peroxiredoxin IV C51A mutant in reduced form | | Descriptor: | Peroxiredoxin-4, SULFATE ION | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
