6O85
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B bound to eukaryotic translation initiation factor 2 from Homo sapiens | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Nguyen, H.C, Kenner, L.R, Frost, A.S. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.
Science, 364, 2019
|
|
6O81
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B bound to translation initiation factor 2 from Homo sapiens | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Nguyen, H, Kenner, L, Frost, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.
Science, 364, 2019
|
|
2IN4
 
 | | Crystal Structure of Myoglobin with Charge Neutralized Heme, ZnDMb-dme | | Descriptor: | Myoglobin, SULFATE ION, ZINC(II)-DEUTEROPORPHYRIN DIMETHYLESTER | | Authors: | Wheeler, K.E, Nocek, J.M, Cull, D.A, Yatsunyk, L.A, Rosenzweig, A.C, Hoffman, B.M. | | Deposit date: | 2006-10-05 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dynamic docking of cytochrome b5 with myoglobin and alpha-hemoglobin: heme-neutralization "squares" and the binding of electron-transfer-reactive configurations.
J.Am.Chem.Soc., 129, 2007
|
|
6CAJ
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B from Homo sapiens | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Tsai, J.C, Miller-Vedam, L.E, Anand, A.A, Jaishankar, P, Nguyen, H.C, Renslo, A.R, Frost, A, Walter, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule.
Science, 359, 2018
|
|
1PV0
 
 | | Structure of the Sda antikinase | | Descriptor: | Sda | | Authors: | Rowland, S.L, Burkholder, W.F, Maciejewski, M.W, Grossman, A.D, King, G.F. | | Deposit date: | 2003-06-26 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of Sda: an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis
Mol.Cell, 13, 2004
|
|
4A6B
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | METHYL ((S)-1-(2-([1,1'-BIPHENYL]-4-YLMETHYL)-2-(3-((3S,4S)-3-BENZYL-4-HYDROXY-1-((1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL)-2-OXOPYRROLIDIN-3-YL)PROPYL)HYDRAZINYL)-3,3-DIMETHYL-1-OXOBUTAN-2-YL)CARBAMATE, POL PROTEIN | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Unge, J, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B.B, Unge, J, Larhed, M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
6OFX
 
 | | Non-rotated ribosome (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6O9Z
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B bound to eukaryotic translation initiation factor 2 from Homo sapiens | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Nguyen, H.C, Kenner, L.R, Frost, A.S. | | Deposit date: | 2019-03-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.
Science, 364, 2019
|
|
6OG7
 
 | | 70S termination complex with RF2 bound to the UGA codon. Non-rotated ribosome with RF2 bound (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6QT9
 
 | | Cryo-EM structure of SH1 full particle. | | Descriptor: | ORF 24, ORF 25, ORF 31, ... | | Authors: | De Colibus, L, Roine, E, Walter, T.S, Ilca, S.L, Wang, X, Wang, N, Roseman, A.M, Bamford, D, Huiskonen, J.T, Stuart, D.I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Assembly of complex viruses exemplified by a halophilic euryarchaeal virus.
Nat Commun, 10, 2019
|
|
1PLS
 
 | | SOLUTION STRUCTURE OF A PLECKSTRIN HOMOLOGY DOMAIN | | Descriptor: | PLECKSTRIN HOMOLOGY DOMAIN | | Authors: | Yoon, H.S, Hajduk, P.J, Petros, A.M, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pleckstrin-homology domain.
Nature, 369, 1994
|
|
4A4Q
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | PROTEASE, methyl [(2S)-1-{2-(2-{(3R,4S)-3-benzyl-4-hydroxy-1-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-2-oxopyrrolidin-3-yl}ethyl)-2-[4-(pyridin-4-yl)benzyl]hydrazinyl}-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B, Unge, J, Larhed, M. | | Deposit date: | 2011-10-19 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
2W0F
 
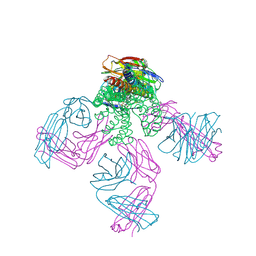 | | Potassium Channel KcsA-Fab Complex with Tetraoctylammonium | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Focia, P.J, Wagner, T, Gross, A. | | Deposit date: | 2008-08-14 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
5ELA
 
 | |
5ICQ
 
 | |
1Y3J
 
 | | Solution structure of the copper(I) form of the fifth domain of Menkes protein | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
2H79
 
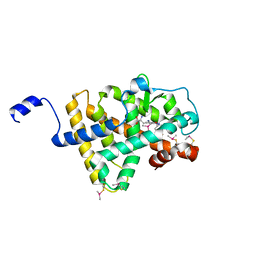 | | Crystal Structure of human TR alpha bound T3 in orthorhombic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
2HC8
 
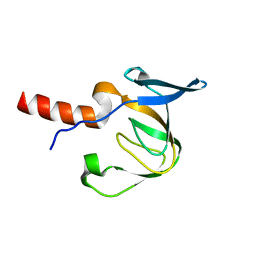 | | Structure of the A. fulgidus CopA A-domain | | Descriptor: | Cation-transporting ATPase, P-type | | Authors: | Sazinsky, M.H, Agawal, S, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-06-15 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Actuator Domain from the Archaeoglobus fulgidus Cu(+)-ATPase(,).
Biochemistry, 45, 2006
|
|
6E1C
 
 | |
1TL5
 
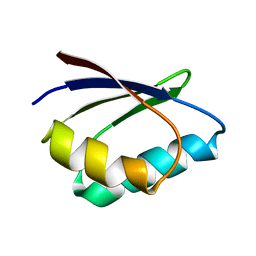 | | Solution structure of apoHAH1 | | Descriptor: | Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
5ICU
 
 | | The crystal structure of CopC from Methylosinus trichosporium OB3b | | Descriptor: | CHLORIDE ION, COPPER (II) ION, CopC, ... | | Authors: | Lawton, T.J, Hurley, J.D, Rosenzweig, A.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The CopC Family: Structural and Bioinformatic Insights into a Diverse Group of Periplasmic Copper Binding Proteins.
Biochemistry, 55, 2016
|
|
4D8G
 
 | | Chlamydia trachomatis NrdB with a Mn/Fe cofactor (procedure 2 - low Mn) | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Dassama, L.M.K, Boal, A.K, Krebs, C, Rosenzweig, A.C, Bollinger Jr, J.M. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1.
J.Am.Chem.Soc., 134, 2012
|
|
2HU9
 
 | | X-ray structure of the Archaeoglobus fulgidus CopZ N-terminal Domain | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, Mercuric transport protein periplasmic component, ... | | Authors: | Sazinsky, M.H, LeMoine, B, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and structure of a Zn2+ and [2Fe-2S]-containing copper chaperone from Archaeoglobus fulgidus.
J.Biol.Chem., 282, 2007
|
|
1N9C
 
 | |
1SMQ
 
 | | Structure of the Ribonucleotide Reductase Rnr2 Homodimer from Saccharomyces cerevisiae | | Descriptor: | Ribonucleoside-diphosphate reductase small chain 1 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
