2AWK
 
 | | GFP R96M mature chromophore | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
3S5D
 
 | |
3S5F
 
 | |
3T3T
 
 | |
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of EV D68 3C protease - to be published
To Be Published
|
|
2A8H
 
 | | Crystal structure of catalytic domain of TACE with Thiomorpholine Sulfonamide Hydroxamate inhibitor | | Descriptor: | 4-({4-[(4-AMINOBUT-2-YNYL)OXY]PHENYL}SULFONYL)-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Jin, G, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-07-08 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acetylenic TACE inhibitors. Part 3: Thiomorpholine sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3URB
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V/R67H | | Descriptor: | COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, IMIDAZOLE, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3T3J
 
 | |
3URA
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V/S61T | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3URN
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V/S61T with cyclohexyl methylphosphonate inhibitor | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3UPM
 
 | | Crystal Structure of PTE mutant H254Q/H257F/K185R/I274N | | Descriptor: | COBALT (II) ION, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3UR5
 
 | | Crystal Structure of PTE mutant K185R/I274N | | Descriptor: | COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3URQ
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V/R67H with cyclohexyl methylphosphonate inhibitor | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3DM0
 
 | | Maltose Binding Protein fusion with RACK1 from A. thaliana | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein fused with RACK1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ullah, H, Scappini, E.L, Moon, A.F, Williams, L.V, Armstrong, D.L, Pedersen, L.C. | | Deposit date: | 2008-06-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana.
Protein Sci., 17, 2008
|
|
3UR2
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, IMIDAZOLE, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3T3X
 
 | |
4BQJ
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
3S4M
 
 | |
3T3L
 
 | |
5A3B
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, ALANINE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
1ZXC
 
 | | Crystal structure of catalytic domain of TNF-alpha converting enzyme (TACE) with inhibitor | | Descriptor: | (3S)-4-{[4-(BUT-2-YNYLOXY)PHENYL]SULFONYL}-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Zhang, Y, Jin, G, Cowling, R, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylenic TACE inhibitors. Part 2: SAR of six-membered cyclic sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3S5E
 
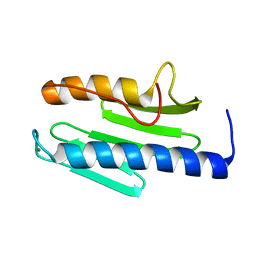 | | Crystal structure of human frataxin variant W155R, one of the Friedreich's ataxia point mutations | | Descriptor: | Frataxin, mitochondrial, MAGNESIUM ION | | Authors: | Tsai, C.-L, Bridwell-Rabb, J, Barondeau, D.P. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Friedreich's Ataxia Variants I154F and W155R Diminish Frataxin-Based Activation of the Iron-Sulfur Cluster Assembly Complex.
Biochemistry, 50, 2011
|
|
3F7K
 
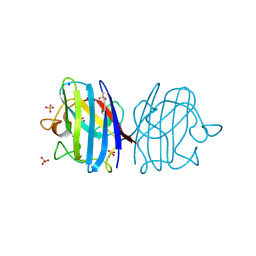 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
3F7L
 
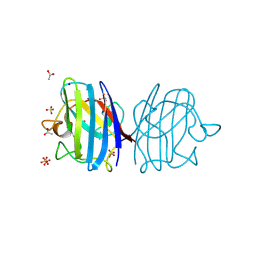 | | X-ray Crystal Structure of Alvinella pompejana Cu,Zn Superoxide Dismutase | | Descriptor: | ACETIC ACID, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
