1WKD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKE
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKF
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1PUD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-06-28 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange.
EMBO J., 15, 1996
|
|
2J49
 
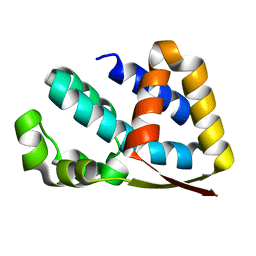 | | Crystal structure of yeast TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 5 | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
2J4B
 
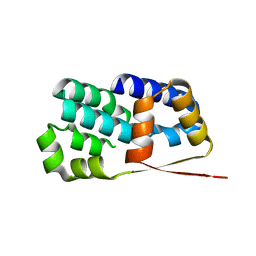 | | Crystal structure of Encephalitozoon cuniculi TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 72/90-100 KDA | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
1AK0
 
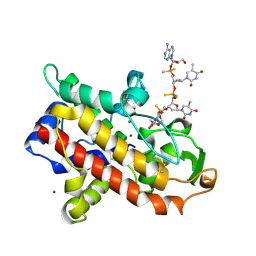 | | P1 NUCLEASE IN COMPLEX WITH A SUBSTRATE ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-(DITHIO)PHOSPHATE, ... | | Authors: | Romier, C, Suck, D. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs.
Proteins, 32, 1998
|
|
1N1J
 
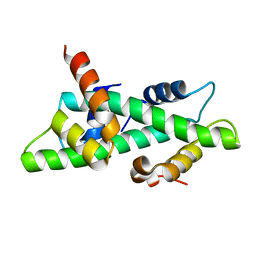 | | Crystal structure of the NF-YB/NF-YC histone pair | | Descriptor: | NF-YB, NF-YC | | Authors: | Romier, C, Cocchiarella, F, Mantovani, R, Moras, D. | | Deposit date: | 2002-10-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y
J.Biol.Chem., 278, 2003
|
|
4ATG
 
 | | TAF6 C-terminal domain from Antonospora locustae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TAF6 | | Authors: | Romier, C. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | TFIID Taf6-Taf9 Complex Formation Involves the Heat Repeat-Containing C-Terminal Domain of Taf6 and is Modulated by Taf5 Protein.
J.Biol.Chem., 287, 2012
|
|
1HLO
 
 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
6TLD
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor 2 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Romier, C. | | Deposit date: | 2019-12-02 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Triazole-Based smHDAC8 Inhibitors.
Chemmedchem, 15, 2020
|
|
7Q1B
 
 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, GLYCEROL, Histone deacetylase DAC2, ... | | Authors: | Marek, M, Ramos-Morales, E, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
7Q1C
 
 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with a hydroxamate inhibitor | | Descriptor: | (E)-3-dibenzofuran-4-yl-N-oxidanyl-prop-2-enamide, Histone deacetylase DAC2, POTASSIUM ION, ... | | Authors: | Ramos-Morales, E, Marek, M, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
8P0A
 
 | | Human Cohesin ATPase module | | Descriptor: | 64-kDa C-terminal product, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Landwerlin, P, Durand, A, Diebold-Durand, M.-L, Romier, C. | | Deposit date: | 2023-05-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Human Cohesin ATPase module
To Be Published
|
|
8PQ5
 
 | | Human Cohesin ATPase module with an open DNA exit gate | | Descriptor: | 64-kDa C-terminal product, ADENOSINE-5'-TRIPHOSPHATE, Structural maintenance of chromosomes protein 1A, ... | | Authors: | Landwerlin, P, Durand, A, Diebold-Durand, M.-L, Romier, C. | | Deposit date: | 2023-07-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Conformational dynamics of the human Cohesin ATPase module
To Be Published
|
|
5FUG
 
 | | Crystal structure of a human YL1-H2A.Z-H2B complex | | Descriptor: | HISTONE H2A.Z, HISTONE H2B TYPE 1-J, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 72 HOMOLOG | | Authors: | Latrick, C.M, Marek, M, Ouararhni, K, Papin, C, Stoll, I, Ignatyeva, M, Obri, A, Ennifar, E, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis and Specificity of H2A.Z-H2B Recognition and Deposition by the Histone Chaperone Yl1
Nat.Struct.Mol.Biol., 23, 2016
|
|
2KKR
 
 | | Solution structure of SCA7 zinc finger domain from human ataxin-7 protein | | Descriptor: | Ataxin-7, ZINC ION | | Authors: | Wang, Y, Atkinson, A.R, Bonnet, J, Romier, C, Kieffer, B, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Histone deubiquitination by SAGA is modulated by an atypical zinc finger domain of Ataxin-7
To be Published
|
|
5FUE
 
 | |
1BH9
 
 | | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE WITH BOUND PCMBS | | Descriptor: | PARA-MERCURY-BENZENESULFONIC ACID, TAFII18, TAFII28 | | Authors: | Birck, C, Poch, O, Romier, C, Ruff, M, Mengus, G, Lavigne, A.-C, Davidson, I, Moras, D. | | Deposit date: | 1998-06-16 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BH8
 
 | | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE | | Descriptor: | TAFII18, TAFII28 | | Authors: | Birck, C, Poch, O, Romier, C, Ruff, M, Mengus, G, Lavigne, A.-C, Davidson, I, Moras, D. | | Deposit date: | 1998-06-16 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family.
Cell(Cambridge,Mass.), 94, 1998
|
|
2XPL
 
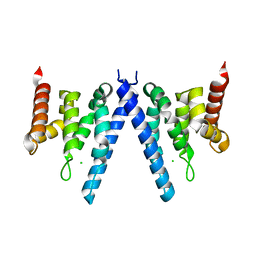 | | Crystal structure of Iws1(Spn1) conserved domain from Encephalitozoon cuniculi | | Descriptor: | CHLORIDE ION, IWS1 | | Authors: | Koch, M, Diebold, M.-L, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
8ROB
 
 | |
8ROF
 
 | |
8RO7
 
 | |
8RO8
 
 | |
