1M6J
 
 | |
5JP5
 
 | | Crystal structure of rat Galectin 5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galectin-5 | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2016-05-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Characterization of Rat Galectin-5, an N-Tailed Monomeric Proto-Type-like Galectin.
Biomolecules, 11, 2021
|
|
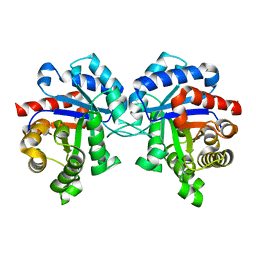
1SUX
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI AND 3-(2-benzothiazolylthio)-1-propanesulfonic acid | | Descriptor: | 3-(2-BENZOTHIAZOLYLTHIO)-1-PROPANESULFONIC ACID, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Tellez-Valencia, A, Olivares-Illana, V, Hernandez-Santoyo, A, Perez-Montfort, R, Costas, M, Rodriguez-Romero, A, Tuena De Gomez-Puyou, M, Gomez-Puyou, A. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface.
J.Mol.Biol., 341, 2004
|
|
3LSE
 
 | | N-Domain of human adhesion/growth-regulatory galectin-9 in complex with lactose | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2010-02-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution.
Int.J.Biochem.Cell Biol., 42, 2010
|
|
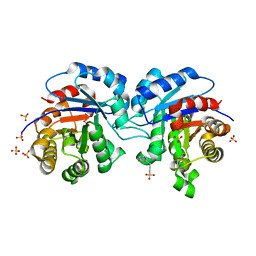
2XUS
 
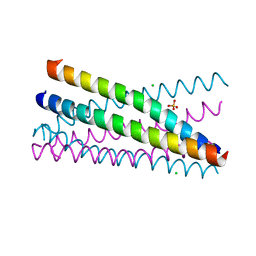 | | Crystal Structure of the BRMS1 N-terminal region | | Descriptor: | BREAST CANCER METASTASIS-SUPPRESSOR 1, CHLORIDE ION, SULFATE ION | | Authors: | Spinola-Amilibia, M, Rivera, J, Ortiz-Lombardia, M, Romero, A, Neira, J.L, Bravo, J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The Structure of Brms1 Nuclear Export Signal and Snx6 Interacting Region Reveals a Hexamer Formed by Antiparallel Coiled Coils.
J.Mol.Biol., 411, 2011
|
|
4GNY
 
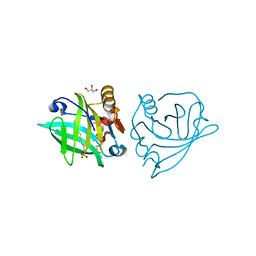 | | Bovine beta-lactoglobulin complex with dodecyl sulfate | | Descriptor: | Beta-lactoglobulin, DODECYL SULFATE, GLYCEROL | | Authors: | Gutierrez-Magdaleno, G, Torres-Rivera, A, Garcia-Hernandez, E, Rodriguez-Romero, A. | | Deposit date: | 2012-08-17 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6367 Å) | | Cite: | Ligand-binding and self-association cooperativity of beta-lactoglobulin
J.Mol.Recognit., 26, 2013
|
|
5VIZ
 
 | |
1WKX
 
 | |
4WVV
 
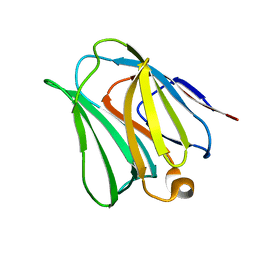 | |
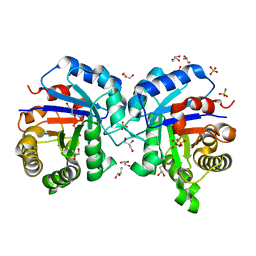
4FF7
 
 | |
3Q3U
 
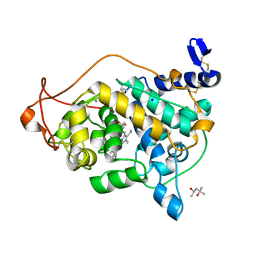 | | Trametes cervina lignin peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Calvino, F.R, Romero, A, Miki, Y, Martinez, A.T. | | Deposit date: | 2010-12-22 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine.
J.Biol.Chem., 286, 2011
|
|
1AZW
 
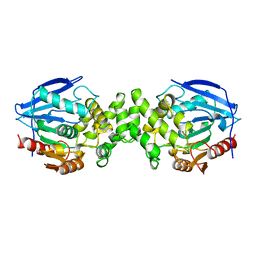 | | PROLINE IMINOPEPTIDASE FROM XANTHOMONAS CAMPESTRIS PV. CITRI | | Descriptor: | PROLINE IMINOPEPTIDASE | | Authors: | Medrano, F.J, Alonso, J, Garcia, J.L, Romero, A, Bode, W, Gomis-Ruth, F.X. | | Deposit date: | 1997-11-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of proline iminopeptidase from Xanthomonas campestris pv. citri: a prototype for the prolyl oligopeptidase family.
EMBO J., 17, 1998
|
|
2JC7
 
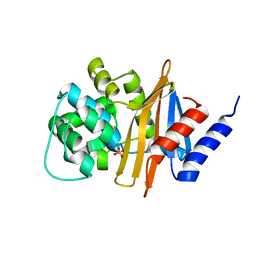 | |
4UWW
 
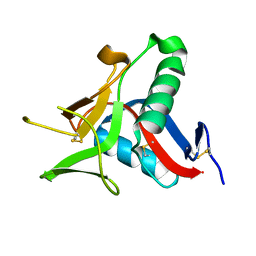 | | Crystallographic Structure of the Intramineral Protein Struthicalcin from Struthio camelus Eggshell | | Descriptor: | STRUTHIOCALCIN-1 | | Authors: | Ruiz, R.R, Moreno, A, Romero, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Structure of Struthiocalcin-1, an Intramineral Protein from Struthio Camelus Eggshell, in Two Different Crystal Forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YMW
 
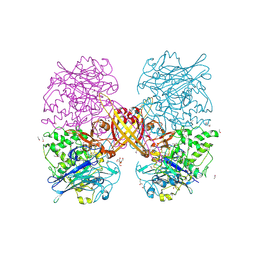 | |
4BIM
 
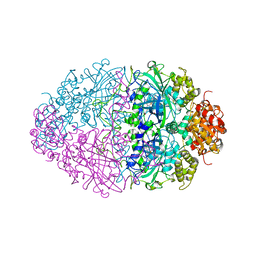 | |
1HKN
 
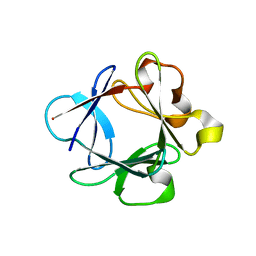 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
4UXM
 
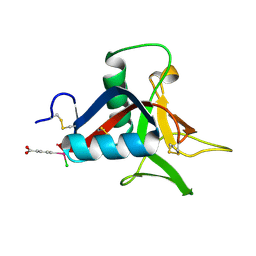 | | Crystal Structure of Struthiocalcin-1, a different crystal form. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, STRUTHIOCALCIN-1 | | Authors: | Ruiz-Arellano, R.R, Moreno, A, Romero, A. | | Deposit date: | 2014-08-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Struthiocalcin-1, an Intramineral Protein from Struthio Camelus Eggshell, in Two Different Crystal Forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WVW
 
 | | Chicken Galectin-8 N-terminal domain complexed with 3'-sialyl-lactose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galectin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2014-11-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combining Crystallography and Hydrogen-Deuterium Exchange to Study Galectin-Ligand Complexes.
Chemistry, 21, 2015
|
|
4AJ9
 
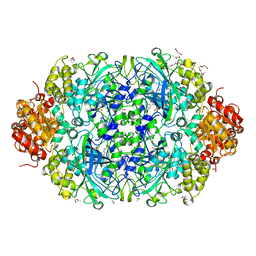 | | Catalase 3 from Neurospora crassa | | Descriptor: | ACETATE ION, CATALASE-3, PENTAETHYLENE GLYCOL, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
7SXD
 
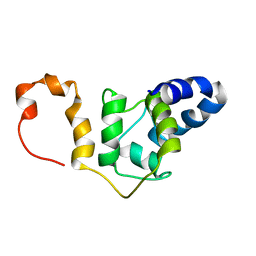 | | NMR solution structure TnC-TnI chimera | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle chimera | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
5IR3
 
 | |
1GZW
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN GALECTIN-1 | | Descriptor: | BETA-MERCAPTOETHANOL, GALECTIN-1, SULFATE ION, ... | | Authors: | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | Deposit date: | 2002-06-07 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding
J.Mol.Biol., 343, 2004
|
|
4W7J
 
 | |
4W7K
 
 | |
