8U37
 
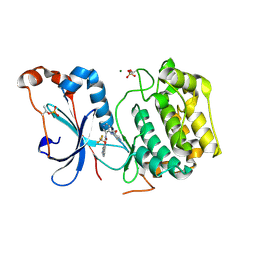 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with NVP-CJL037 at 2.48-A resolution | | Descriptor: | (6M)-3-amino-N-{4-[(3R,4S)-4-amino-3-methoxypiperidin-1-yl]pyridin-3-yl}-6-[3-(trifluoromethoxy)pyridin-2-yl]pyrazine-2-carboxamide, MAGNESIUM ION, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8UAK
 
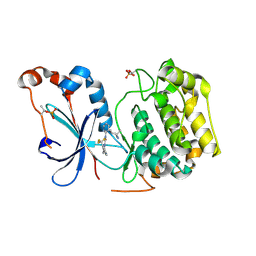 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
5LKW
 
 | |
4OKJ
 
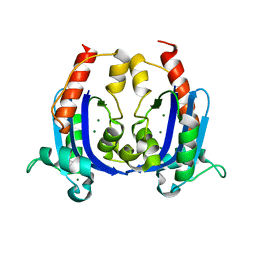 | | Crystal structure of RNase AS from M tuberculosis | | Descriptor: | 3'-5' exoribonuclease Rv2179c/MT2234.1, MAGNESIUM ION | | Authors: | Romano, M, van de Weerd, R, Brouwer, F.C.C, Roviello, G.N, Lacroix, R, Sparrius, M, van den Brink-van Stempvoort, G, Maaskant, J.J, van der Sar, A.M, Appelmelk, B.J, Geurtsen, J.J, Berisio, R. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of RNase AS, a Polyadenylate-Specific Exoribonuclease Affecting Mycobacterial Virulence In Vivo
Structure, 22, 2014
|
|
4OKK
 
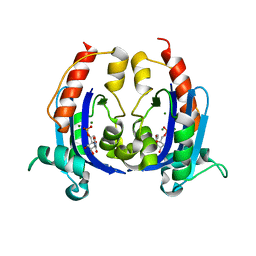 | | Crystal structure of RNase AS from M tuberculosis in complex with UMP | | Descriptor: | 3'-5' exoribonuclease Rv2179c/MT2234.1, MAGNESIUM ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Romano, M, van de Weerd, R, Brouwer, F.C.C, Roviello, G.N, Lacroix, R, Sparrius, M, van den Brink-van Stempvoort, G, Maaskant, J.J, van der Sar, A.M, Appelmelk, B.J, Geurtsen, J.J, Berisio, R. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Function of RNase AS, a Polyadenylate-Specific Exoribonuclease Affecting Mycobacterial Virulence In Vivo
Structure, 22, 2014
|
|
1RWO
 
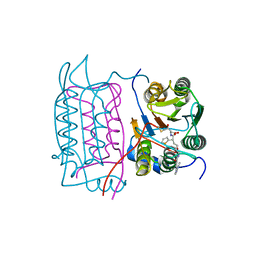 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-{6-[4-(quinoxalin-2-ylamino)-benzoylamino]-2-thiophen-2-yl-hexanoylamino}-pentanoic acid | | Descriptor: | 4-OXO-3-{6-[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-2-THIOPHEN-2-YL-HEXANOYLAMINO}-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1JSX
 
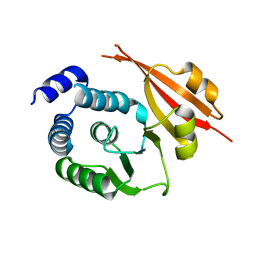 | |
1RWP
 
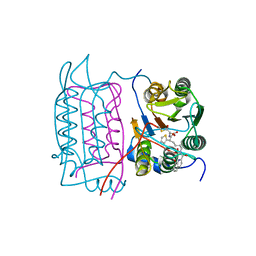 | |
1RWX
 
 | |
1SC4
 
 | |
1RWN
 
 | |
1RWM
 
 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-[2-(5-{[4-(quinoxalin-2-ylamino)-benzoylamino]-methyl}-thiophen-2-yl)-acetylamino]-pentanoic acid | | Descriptor: | 4-OXO-3-[2-(5-{[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-METHYL}-THIOPHEN-2-YL)-ACETYLAMINO]-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1RWV
 
 | |
1RWW
 
 | |
1SC1
 
 | | Crystal structure of an active-site ligand-free form of the human caspase-1 C285A mutant | | Descriptor: | CHLORIDE ION, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Scheer, J.M, O'Brien, T, McDowell, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding.
Structure, 12, 2004
|
|
1RWK
 
 | | Crystal structure of human caspase-1 in complex with 3-(2-mercapto-acetylamino)-4-oxo-pentanoic acid | | Descriptor: | 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Lam, J.W, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1SC3
 
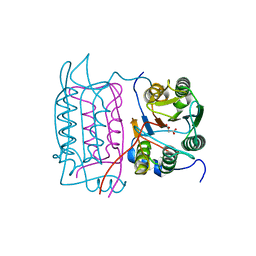 | | Crystal structure of the human caspase-1 C285A mutant in complex with malonate | | Descriptor: | Interleukin-1 beta convertase, MALONATE ION | | Authors: | Romanowski, M.J, Scheer, J.M, O'Brien, T, McDowell, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding.
Structure, 12, 2004
|
|
1JSS
 
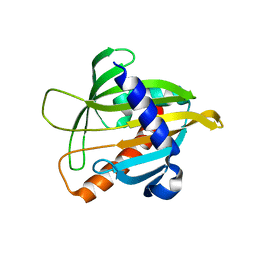 | | Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4). | | Descriptor: | cholesterol-regulated START protein 4 | | Authors: | Romanowski, M.J, Soccio, R.E, Breslow, J.L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-08-17 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1XTY
 
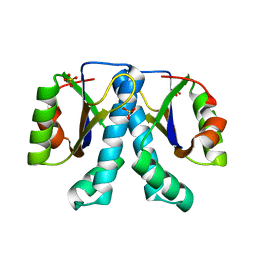 | | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Fromant, M, Schmitt, E, Mechulam, Y, Lazennec, C, Plateau, P, Blanquet, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.
Biochemistry, 44, 2005
|
|
1KAG
 
 | |
1K47
 
 | |
1JYH
 
 | |
8QE4
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 2024
|
|
8QOR
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(CFL)P*(FRG)P*(CFL)P*(FRG)P*(CFL)P*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 2024
|
|
8QDU
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 2024
|
|
