1USV
 
 | |
1UV5
 
 | | GLYCOGEN SYNTHASE KINASE 3 BETA COMPLEXED WITH 6-BROMOINDIRUBIN-3'-OXIME | | Descriptor: | 6-BROMOINDIRUBIN-3'-OXIME, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Dajani, R, Pearl, L.H, Roe, S.M. | | Deposit date: | 2004-01-14 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gsk-3-Selective Inhibitors Derived from Tyrian Purple Indurubins
Chem.Biol., 10, 2003
|
|
1UNN
 
 | |
1USU
 
 | |
1UWJ
 
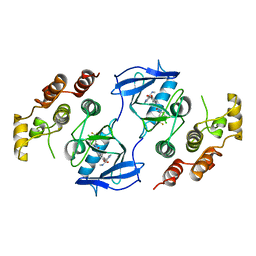 | | The complex of mutant V599E B-RAF and BAY439006. | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Barford, D, Roe, S.M, Wan, P.T.C, Cancer Genome Project | | Deposit date: | 2004-02-05 | | Release date: | 2004-03-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Mechanism of Activation of the Raf-Erk Signaling Pathway by Oncogenic Mutations of B-Raf
Cell(Cambridge,Mass.), 116, 2004
|
|
1W4S
 
 | |
1O9U
 
 | |
1ODG
 
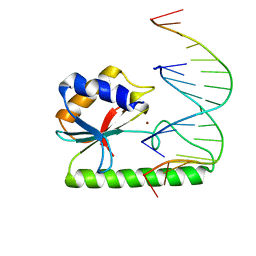 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
1AM1
 
 | |
1AMW
 
 | |
1QO0
 
 | |
1QNL
 
 | |
2BGG
 
 | | The structure of a Piwi protein from Archaeoglobus fulgidus complexed with a 16nt siRNA duplex. | | Descriptor: | 5'-R(*GP*UP*CP*GP*AP*AP*UP*UP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*GP*CP)-3', MANGANESE (II) ION, ... | | Authors: | Parker, J.S, Roe, S.M, Barford, D. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Mrna Recognition from a Piwi Domain-Sirna Guide Complex
Nature, 434, 2005
|
|
2XX4
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (E)-ETHYL 13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDROBENZO[C][1]AZACYCLOTETRADECINE-10-CARBOXYLATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
2XX5
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (5E,10R)-N-BENZYL-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDRO-2-BENZAZACYCLOTETRADECINE-10-CARBOXAMIDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
2XX2
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (5E)-13-CHLORO-14,16-DIHYDROXY-3,4,7,8,9,10-HEXAHYDRO-2-BENZAZACYCLOTETRADECINE-1,11(2H,12H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
2V7E
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 (CARM1), unliganded | | Descriptor: | HISTONE-ARGININE METHYLTRANSFERASE CARM1, MERCURY (II) ION | | Authors: | Yue, W.W, Hassler, M, Roe, S.M, Thompson-Vale, V, Pearl, L.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights Into Histone Code Syntax from Structural and Biochemical Studies of Carm1 Methyltransferase
Embo J., 26, 2007
|
|
2V74
 
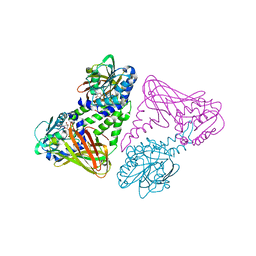 | | Crystal structure of coactivator-associated arginine methyltransferase 1 (CARM1), in complex with S-adenosyl-homocysteine | | Descriptor: | HISTONE-ARGININE METHYLTRANSFERASE CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yue, W.W, Hassler, M, Roe, S.M, Thompson-Vale, V, Pearl, L.H. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights Into Histone Code Syntax from Structural and Biochemical Studies of Carm1 Methyltransferase
Embo J., 26, 2007
|
|
2W2W
 
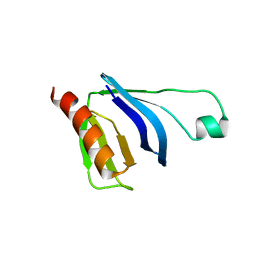 | | PLCg2 Split Pleckstrin Homology (PH) Domain | | Descriptor: | 1-PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE PHOSPHODIESTERASE GAMMA-2 | | Authors: | Opaleye, O, Bunney, T.D, Roe, S.M, Pearl, L.H. | | Deposit date: | 2008-11-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Formation of an Active Signaling Complex between Rac and Phospholipase C Gamma 2.
Mol.Cell, 34, 2009
|
|
2W42
 
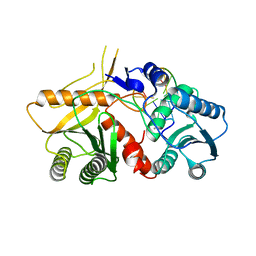 | | THE STRUCTURE OF A PIWI PROTEIN FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH A 16NT DNA DUPLEX. | | Descriptor: | 5'-D(*GP*TP*CP*GP*AP*AP*TP*TP)-3', 5'-D(*TP*TP*CP*GP*AP*CP*GP*CP)-3', MANGANESE (II) ION, ... | | Authors: | Parker, J.S, Roe, S.M, Barford, D. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enhancement of the Seed-Target Recognition Step in RNA Silencing by a Piwi-Mid Domain Protein
Mol.Cell, 33, 2009
|
|
2VXC
 
 | |
2WMO
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WT8
 
 | |
2WMN
 
 | | Structure of the complex between DOCK9 and Cdc42-GDP. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2W2V
 
 | | Rac2 (G12V) in complex with GTPgS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 2 | | Authors: | Opaleye, O, Bunney, T.D, Roe, S.M, Pearl, L.H. | | Deposit date: | 2008-11-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Formation of an Active Signaling Complex between Rac and Phospholipase C Gamma 2.
Mol.Cell, 34, 2009
|
|
