1HMV
 
 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2O36
 
 | |
2O3E
 
 | |
4P08
 
 | | Engineered thermostable dimeric cocaine esterase | | Descriptor: | Cocaine esterase | | Authors: | Rodgers, D.W, Chow, K.-M, Fang, L, Zhan, C.-G. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Rational design, preparation, and characterization of a therapeutic enzyme mutant with improved stability and function for cocaine detoxification.
Acs Chem.Biol., 9, 2014
|
|
8SW1
 
 | |
8SW0
 
 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
3TUV
 
 | |
4FXY
 
 | | Crystal structure of rat neurolysin with bound pyrazolidin inhibitor | | Descriptor: | 1-{(2S)-1-[(3R)-3-(2-chlorophenyl)-2-(2-fluorophenyl)pyrazolidin-1-yl]-1-oxopropan-2-yl}-3-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]urea, Neurolysin, mitochondrial, ... | | Authors: | Rodgers, D.W, Hines, C.S. | | Deposit date: | 2012-07-03 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of the neuropeptidase neurolysin.
J.Biol.Chem., 289, 2014
|
|
3CPM
 
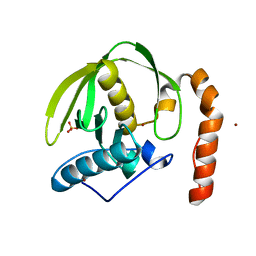 | | plant peptide deformylase PDF1B crystal structure | | Descriptor: | Peptide deformylase, chloroplast, SULFATE ION, ... | | Authors: | Rodgers, D.W, Houtz, R.L, Dirk, L.M.A, Schmidt, J.J, Cai, Y. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture
Biochem.J., 413, 2008
|
|
1PER
 
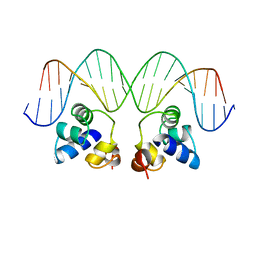 | |
3P7O
 
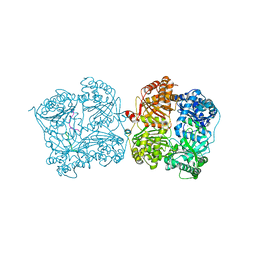 | |
3P7L
 
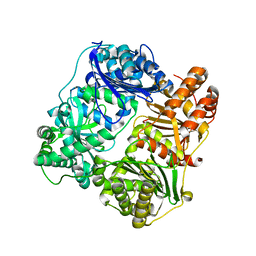 | |
1Q6X
 
 | | Crystal structure of rat choline acetyltransferase | | Descriptor: | SODIUM ION, choline O-acetyltransferase | | Authors: | Cai, Y, Rodgers, D.W. | | Deposit date: | 2003-08-14 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Choline acetyltransferase structure reveals distribution of mutations that cause motor disorders.
Embo J., 23, 2004
|
|
5J8D
 
 | | Structure of nitroreductase from E. cloacae complexed with nicotinic acid adenine dinucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID ADENINE DINUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
5J8G
 
 | | Structure of nitroreductase from E. cloacae complexed with para-nitrobenzoic acid | | Descriptor: | 4-NITROBENZOIC ACID, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.-F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
1S4B
 
 | | Crystal structure of human thimet oligopeptidase. | | Descriptor: | Thimet oligopeptidase, ZINC ION | | Authors: | Ray, K, Hines, C.S, Coll-Rodriguez, J, Rodgers, D.W. | | Deposit date: | 2004-01-15 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization
J.Biol.Chem., 279, 2004
|
|
2OR1
 
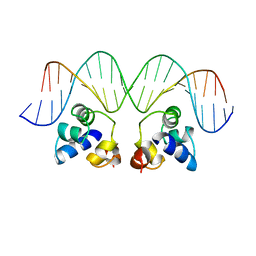 | | RECOGNITION OF A DNA OPERATOR BY THE REPRESSOR OF PHAGE 434. A VIEW AT HIGH RESOLUTION | | Descriptor: | 434 REPRESSOR, DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3') | | Authors: | Aggarwal, A.K, Rodgers, D.W, Drottar, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1989-09-05 | | Release date: | 1989-09-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a DNA operator by the repressor of phage 434: a view at high resolution.
Science, 242, 1988
|
|
2BKB
 
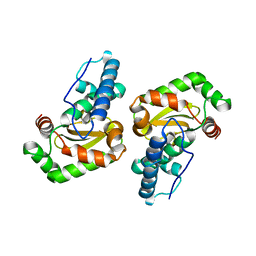 | | q69e-FeSOD | | Descriptor: | FE (II) ION, SUPEROXIDE DISMUTASE [FE] | | Authors: | Yikilmaz, E, Rodgers, D.W, Miller, A.-F. | | Deposit date: | 2005-02-14 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crucial Importance of Chemistry in the Structure-Function Link: Manipulating Hydrogen Bonding in Iron-Containing Superoxide Dismutase.
Biochemistry, 45, 2006
|
|
1ZA5
 
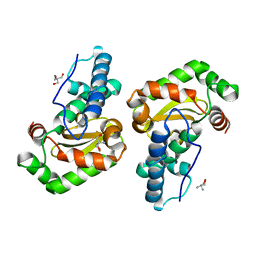 | | Q69H-FeSOD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, Superoxide dismutase [Fe] | | Authors: | Yikilmaz, E, Rodgers, D.W, Miller, A.F. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crucial Importance of Chemistry in the Structure-Function Link: Manipulating Hydrogen Bonding in Iron-Containing Superoxide Dismutase.
Biochemistry, 45, 2006
|
|
1I1I
 
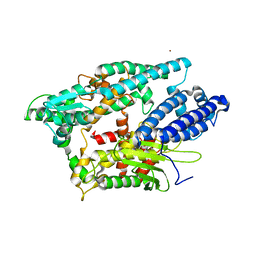 | | NEUROLYSIN (ENDOPEPTIDASE 24.16) CRYSTAL STRUCTURE | | Descriptor: | NEUROLYSIN, ZINC ION | | Authors: | Brown, C.K, Madauss, K, Lian, W, Tolbert, W.D, Beck, M.R, Rodgers, D.W. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of neurolysin reveals a deep channel that limits substrate access.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQD
 
 | | Structure of Nitroreductase from E. cloacae Bound with 2e-Reduced Flavin Mononucleotide (FMN) | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQB
 
 | | Structure of Nitroreductase from E. cloacae complex with inhibitor benzoate | | Descriptor: | BENZOIC ACID, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
