7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
6NJT
 
 | | Mouse endonuclease G mutant - H97A | | Descriptor: | CHLORIDE ION, Endonuclease G, mitochondrial, ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
6NJU
 
 | | Mouse endonuclease G mutant H97A bound to A-DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DNA (5'-D(CCGGCGCCGG)-3'), ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
7NBA
 
 | | Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin motor domain-containing protein,Kinesin motor domain-containing protein, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
7NB8
 
 | | Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-5, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
6U0W
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K133M at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-08-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6U0X
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS Q123D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-08-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | Descriptor: | De novo designed transmembrane beta-barrel TMB2.3 | | Authors: | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
6X9Z
 
 | |
6YDL
 
 | | Substrate-free beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6YDM
 
 | | beta-phosphoglucomutase from Lactococcus lactis with citrate, tris and acetate bound | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
5M5C
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5M50
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6YDJ
 
 | | P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with glucose 6-phosphate and trifluoromagnesate | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6YDK
 
 | | Substrate-free P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
5M54
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7NAY
 
 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
7TFF
 
 | | Crystal structure of human platelet phosphofructokinase-1 mutant- D564N | | Descriptor: | ATP-dependent 6-phosphofructokinase, platelet type, PHOSPHATE ION, ... | | Authors: | Hansen, H, Webb, B.A, Robart, A.R, Narayanasami, S. | | Deposit date: | 2022-01-06 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cancer-associated somatic mutations in human phosphofructokinase-1 reveal a critical electrostatic interaction for allosteric regulation of enzyme activity.
Biochem.J., 480, 2023
|
|
2WOU
 
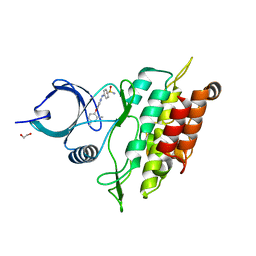 | | ALK5 IN COMPLEX WITH 4-((4-((2,6-dimethyl-3-pyridyl)oxy)-2-pyridyl) amino)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2,6-DIMETHYLPYRIDIN-3-YL)OXY]PYRIDIN-2-YL}AMINO)BENZENESULFONAMIDE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Debreczeni, J.E, Norman, R.A, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
7R18
 
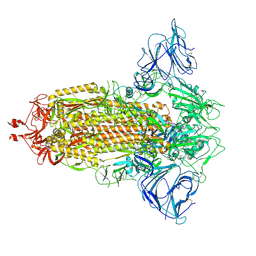 | | Mink Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R14
 
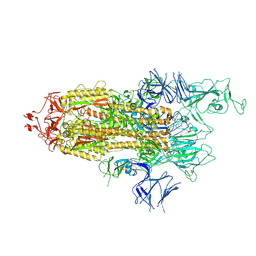 | | Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
2WOT
 
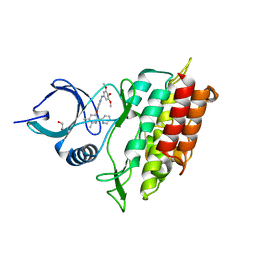 | | ALK5 IN COMPLEX WITH 4-((5,6-dimethyl-2-(2-pyridyl)-3-pyridyl)oxy)-N-(3,4,5-trimethoxyphenyl)pyridin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5,6-DIMETHYL-2,2'-BIPYRIDIN-3-YL)OXY]-N-(3,4,5-TRIMETHOXYPHENYL)PYRIDIN-2-AMINE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Norman, R.A, Debreczeni, J.E, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
7BNN
 
 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R15
 
 | | Alpha Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
