6XHH
 
 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | Descriptor: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XHG
 
 | | Far-red absorbing dark state of JSC1_58120g3 with bound biliverdin IXa (BV) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(~{Z})-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, JSC1_58120g3 | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3EJ1
 
 | | CDK2/CyclinA complexed with a pyrazolopyridazine inhibitor | | Descriptor: | Cell division protein kinase 2, Cyclin-A2, N-cyclopropyl-4-pyrazolo[1,5-b]pyridazin-3-ylpyrimidin-2-amine | | Authors: | Stevens, K, Reno, M, Alberti, J, Price, D, Kane-Carson, L, Knick, V, Shewchuk, L, Hassell, A, Veal, J, Peel, M. | | Deposit date: | 2008-09-17 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Synthesis and evaluation of pyrazolo[1,5-b]pyridazines as selective cyclin dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3EID
 
 | | CDK2/CyclinA complexed with a pyrazolopyridazine inhibitor | | Descriptor: | (2S)-1-(dimethylamino)-3-(4-{[4-(6-morpholin-4-ylpyrazolo[1,5-b]pyridazin-3-yl)pyrimidin-2-yl]amino}phenoxy)propan-2-ol, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Steven, K, Reno, M, Alberti, J, Price, D, Kane-Carson, L, Knick, V, Shewchuk, L, Hassell, A, Veal, J, Peel, M. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and evaluation of pyrazolo[1,5-b]pyridazines as selective cyclin dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6GWS
 
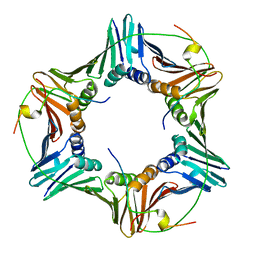 | | Crystal structure of human PCNA in complex with three p15 peptides | | Descriptor: | PCNA-associated factor, Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Gonzalez-Magana, A, Romano-Moreno, M, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
5OSI
 
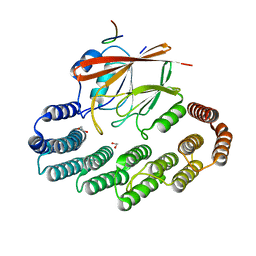 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL harpin loop (163-176) | | Descriptor: | 1,2-ETHANEDIOL, Interaptin, SODIUM ION, ... | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OSH
 
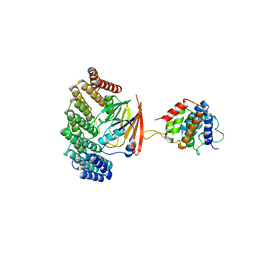 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OT4
 
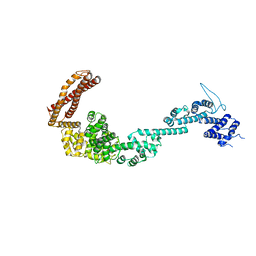 | | Structure of the Legionella pneumophila effector RidL (1-866) | | Descriptor: | GLYCEROL, Interaptin | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3ZDT
 
 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8RKS
 
 | | Structure of VPS29-VPS35 bound to the LFa motif R21 of Fam21. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35, WASH complex subunit 2A | | Authors: | Romano-Moreno, M, Astorga-Simon, E.N, Rojas, A.L, Hierro, A. | | Deposit date: | 2023-12-30 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Protein Sci., 33, 2024
|
|
2VRJ
 
 | | Beta-glucosidase from Thermotoga maritima in complex with N-octyl-5- deoxy-6-oxa-N-(thio)carbamoylcalystegine | | Descriptor: | (1S,2R,3S,4R,5R)-2,3,4-trihydroxy-N-octyl-6-oxa-8-azabicyclo[3.2.1]octane-8-carbothioamide, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Aguilar, M, Gloster, T.M, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Llebaria, A, Casas, J, Egido-Gabas, M, Garcia Fernandez, J.M. | | Deposit date: | 2008-04-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Beta-Glucosidase Inhibition by Ring-Modified Calystegine Analogues.
Chembiochem, 9, 2008
|
|
2WC4
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-thia-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
2WBG
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
2WC3
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-8-epi-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8S,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
2WCG
 
 | | X-ray structure of acid-beta-glucosidase with N-octyl(cyclic guanidine)-nojirimycin in the active site | | Descriptor: | CHLORIDE ION, GLUCOSYLCERAMIDASE, N-[(3E,5R,6R,7S,8R,8AR)-5,6,7,8-TETRAHYDROXYHEXAHYDROIMIDAZO[1,5-A]PYRIDIN-3(2H)-YLIDENE]OCTAN-1-AMINIUM, ... | | Authors: | Brumshtein, B, Aguilar, M, Garcia-Moreno, M.I, Mellet, C.O, Garcia-Fernandez, J.M, Silman, I, Shaaltiel, Y, Aviezer, D, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 6-Amino-6-Deoxy-5,6-Di-N-(N'-Octyliminomethylidene)Nojirimycin: Synthesis, Biological Evaluation, and Crystal Structure in Complex with Acid Beta-Glucosidase.
Chembiochem, 10, 2009
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
4CYE
 
 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6HVO
 
 | | Crystal structure of human PCNA in complex with three peptides of p12 subunit of human polymerase delta | | Descriptor: | DNA polymerase delta subunit 4, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Gonzalez-Magana, A, Romano-Moreno, M, Rojas, A.L, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-23 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The p12 subunit of human polymerase delta uses an atypical PIP box for molecular recognition of proliferating cell nuclear antigen (PCNA).
J.Biol.Chem., 294, 2019
|
|
7Z79
 
 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
4JIX
 
 | | Crystal structure of the metallopeptidase zymogen of Methanocaldococcus jannaschii jannalysin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
4JIU
 
 | | Crystal structure of the metallopeptidase zymogen of Pyrococcus abyssi abylysin | | Descriptor: | GLYCEROL, Proabylysin, ZINC ION | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
1ANX
 
 | |
1OEJ
 
 | | YodA from Escherichia coli crystallised with no added ions | | Descriptor: | HYPOTHETICAL PROTEIN YODA, NICKEL (II) ION | | Authors: | David, G, Blondeau, K, Renouard, M, Penel, S, Lewit-Bentley, A. | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Yoda from Escherichia Coli is a Metal-Binding, Lipocalin-Like Protein
J.Biol.Chem., 278, 2003
|
|
1BT6
 
 | | P11 (S100A10), LIGAND OF ANNEXIN II IN COMPLEX WITH ANNEXIN II N-TERMINUS | | Descriptor: | ANNEXIN II, S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-09-02 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
