9EVV
 
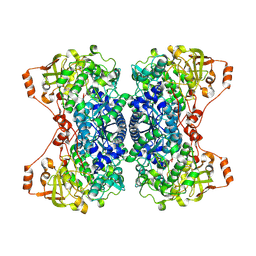 | | His579Leu variant of L-arabinonate dehydratase co-crystallized with 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, FE2/S2 (INORGANIC) CLUSTER, L-arabinonate dehydratase, ... | | Authors: | Ren, Y, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Unveiling the importance of the C-terminus in the sugar acid dehydratase of the IlvD/EDD superfamily.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
6LYG
 
 | |
7DSC
 
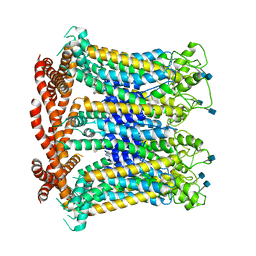
 | | CALHM1 open state with disordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
7DSD
 
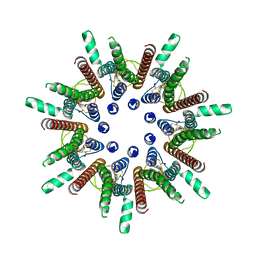 | | CALHM1 close state with disordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
7DSE
 
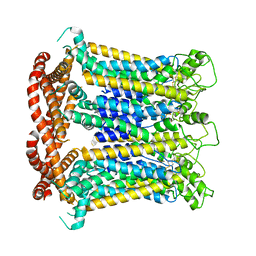 | | CALHM1 close state with ordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
4OWR
 
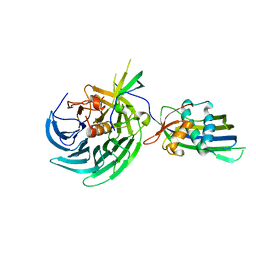 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3K8P
 
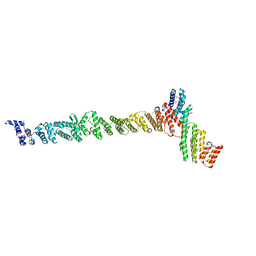 | |
3ETU
 
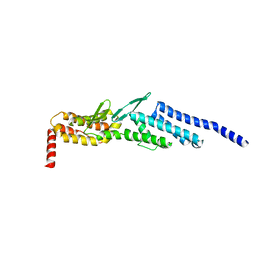 | | Crystal structure of yeast Dsl1p | | Descriptor: | Protein transport protein DSL1 | | Authors: | Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3ETV
 
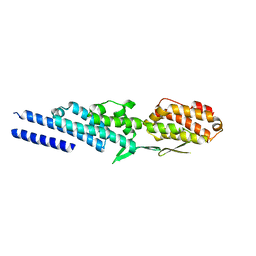 | | Crystal structure of a Tip20p-Dsl1p fusion protein | | Descriptor: | Protein transport protein TIP20, Protein transport protein DSL1 chimera | | Authors: | Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5SUQ
 
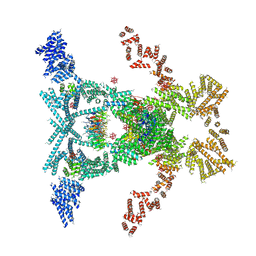 | | Crystal structure of the THO-Sub2 complex | | Descriptor: | 12-TUNGSTOPHOSPHATE, ATP-dependent RNA helicase SUB2, Tex1, ... | | Authors: | Ren, Y, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
5SUP
 
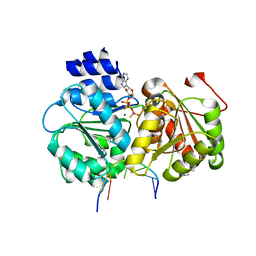 | | Crystal structure of the Sub2-Yra1 complex in association with RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase SUB2, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ren, Y, Schmiege, P, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
2JSD
 
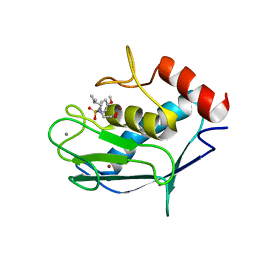 | | Solution structure of MMP20 complexed with NNGH | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-03 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
8XVT
 
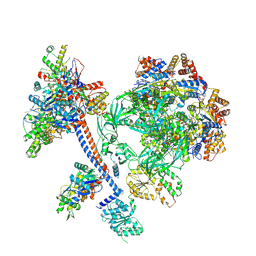 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVG
 
 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XN9
 
 | | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6 | | Descriptor: | 1D6 VH, 1D6 VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, P.F, Ren, Y, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6
To Be Published
|
|
8XNH
 
 | | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 5C8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5C8-VH, 5C8-VL, ... | | Authors: | Fan, P.F, Ren, Y, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6
To Be Published
|
|
8ENK
 
 | |
7YVA
 
 | | Crystal structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with lipoic acid | | Descriptor: | 1,2-ETHANEDIOL, Candida albicans Fructose-1,6-bisphosphate aldolase, LIPOIC ACID, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Crystal structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with lipoic acid
To Be Published
|
|
7V6G
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
7V6F
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
6X25
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7LUV
 
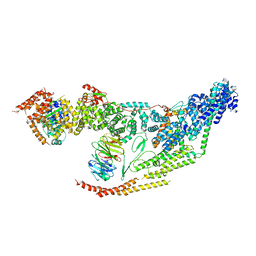 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
6LNK
 
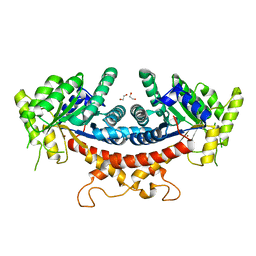 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Huang, Y, Cao, H, Ren, Y, Wan, J. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
8WAS
 
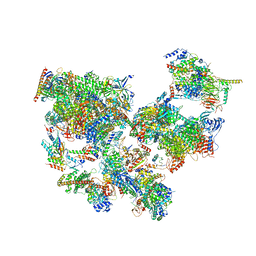 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
