4KE9
 
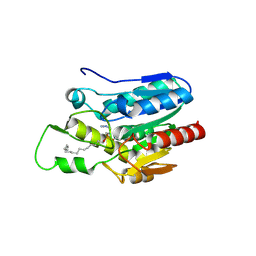 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-stearyol glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, hexadecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3J9Y
 
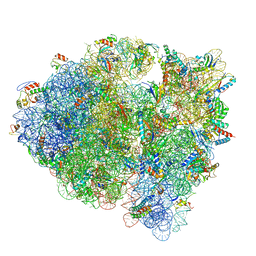 | | Cryo-EM structure of tetracycline resistance protein TetM bound to a translating E.coli ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Nguyen, F, Beckmann, R, Wilson, D.N. | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-15 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9- angstrom resolution.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3J5L
 
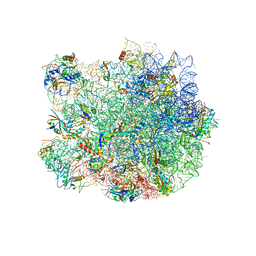 | | Structure of the E. coli 50S subunit with ErmBL nascent chain | | Descriptor: | 23S ribsomal RNA, 5'-R(*CP*(MA6))-3', 5'-R(*CP*CP*A)-3', ... | | Authors: | Arenz, S, Ramu, H, Gupta, P, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Mankin, A.S, Wilson, D.N. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide.
Nat Commun, 5, 2014
|
|
2V8G
 
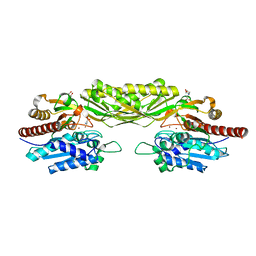 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with the product beta-alanine | | Descriptor: | BETA-ALANINE, BETA-ALANINE SYNTHASE, BICINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8H
 
 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri in complex with its substrate N-carbamyl-beta- alanine | | Descriptor: | BETA-ALANINE SYNTHASE, BICINE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8V
 
 | | Crystal structure of mutant R322A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-ALANINE SYNTHASE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-15 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8D
 
 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-ALANINE SYNTHASE, ZINC ION | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2VZG
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD2 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
2VHH
 
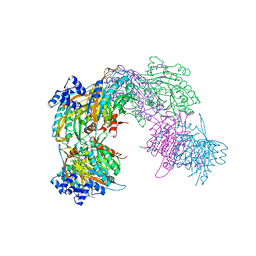 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
1XTU
 
 | | Sulfolobus solfataricus uracil phosphoribosyltransferase in complex with uridine 5'-monophosphate (UMP) and cytidine 5'-triphosphate (CTP) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Probable uracil phosphoribosyltransferase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Arent, S, Harris, P, Jensen, K.F, Larsen, S. | | Deposit date: | 2004-10-24 | | Release date: | 2005-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Regulation and Communication between Subunits in Uracil Phosphoribosyltransferase from Sulfolobus solfataricus(,)
Biochemistry, 44, 2005
|
|
2VZC
 
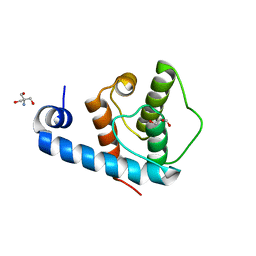 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
2VZI
 
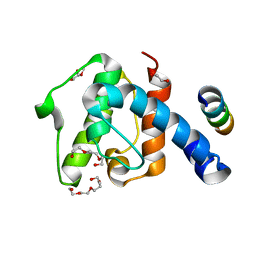 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD4 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin,Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
2VZD
 
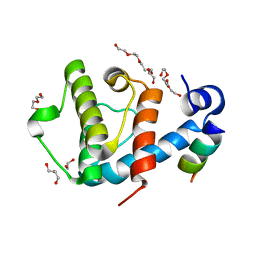 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin in complex with paxillin LD1 motif | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-PARVIN, GLYCEROL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
1XTT
 
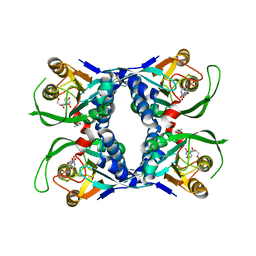 | | Sulfolobus solfataricus uracil phosphoribosyltransferase in complex with uridine 5'-monophosphate (UMP) | | Descriptor: | ACETIC ACID, Probable uracil phosphoribosyltransferase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Arent, S, Harris, P, Jensen, K.F, Larsen, S. | | Deposit date: | 2004-10-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric Regulation and Communication between Subunits in Uracil Phosphoribosyltransferase from Sulfolobus solfataricus(,)
Biochemistry, 44, 2005
|
|
1XTV
 
 | | Sulfolobus solfataricus uracil phosphoribosyltransferase with uridine 5'-monophosphate (UMP) bound to half of the subunits | | Descriptor: | Probable uracil phosphoribosyltransferase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Arent, S, Harris, P, Jensen, K.F, Larsen, S. | | Deposit date: | 2004-10-24 | | Release date: | 2005-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric Regulation and Communication between Subunits in Uracil Phosphoribosyltransferase from Sulfolobus solfataricus(,)
Biochemistry, 44, 2005
|
|
2VHI
 
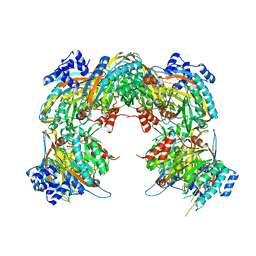 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
2WTB
 
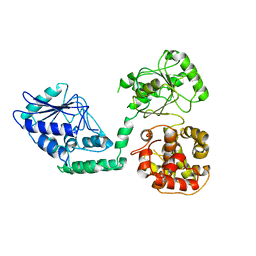 | | Arabidopsis thaliana multifuctional protein, MFP2 | | Descriptor: | FATTY ACID MULTIFUNCTIONAL PROTEIN (ATMFP2) | | Authors: | Arent, S, Pye, V.E, Christensen, C.E, Norgaard, A, Henriksen, A. | | Deposit date: | 2009-09-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Multi-Functional Protein in Peroxisomal Beta-Oxidation. Structure and Substrate Specificity of the Arabidopsis Thaliana Protein, Mfp2
J.Biol.Chem., 285, 2010
|
|
2FXV
 
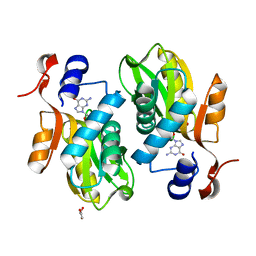 | | Bacillus subtilis Xanthine Phosphoribosyltransferase in Complex with Guanosine 5'-monophosphate (GMP) | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Xanthine phosphoribosyltransferase | | Authors: | Arent, S, Kadziola, A, Larsen, S, Neuhard, J, Jensen, K.F. | | Deposit date: | 2006-02-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Extraordinary Specificity of Xanthine Phosphoribosyltransferase from Bacillus subtilis Elucidated
by Reaction Kinetics, Ligand Binding, and Crystallography
Biochemistry, 45, 2006
|
|
8RAN
 
 | |
8RAO
 
 | |
8RAM
 
 | |
8RAP
 
 | |
1E8T
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8U
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1R43
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri (selenomethionine substituted protein) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-AMINO ISOBUTYRATE, ... | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-03 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278
|
|
