2DUB
 
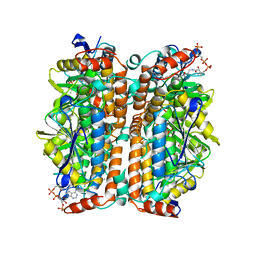 | | ENOYL-COA HYDRATASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | 2-ENOYL-COA HYDRATASE, OCTANOYL-COENZYME A | | Authors: | Engel, C.K, Wierenga, R.K. | | Deposit date: | 1997-04-28 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule.
J.Mol.Biol., 275, 1998
|
|
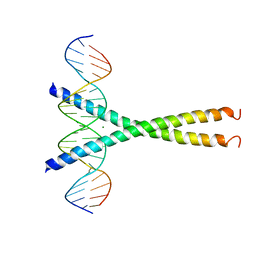
1DH3
 
 | |
1DLU
 
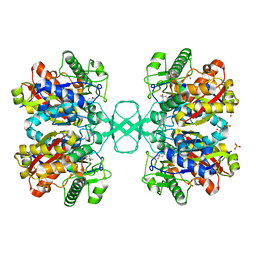 | | UNLIGANDED BIOSYNTHETIC THIOLASE FROM ZOOGLOEA RAMIGERA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIOSYNTHETIC THIOLASE, SULFATE ION | | Authors: | Modis, Y, Wierenga, R.K. | | Deposit date: | 1999-12-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
J.Mol.Biol., 297, 2000
|
|
1DM3
 
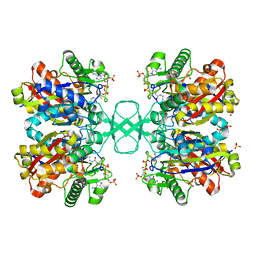 | |
1WET
 
 | | STRUCTURE OF THE PURR-GUANINE-PURF OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Glasfeld, A, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-04-27 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity.
J.Biol.Chem., 272, 1997
|
|
1BD3
 
 | | STRUCTURE OF THE APO URACIL PHOSPHORIBOSYLTRANSFERASE, 2 MUTANT C128V | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
2V2H
 
 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed-Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3D70
 
 | |
1BOW
 
 | | MULTIDRUG-BINDING DOMAIN OF TRANSCRIPTION ACTIVATOR BMRR (APO FORM) | | Descriptor: | MANGANESE (II) ION, MULTIDRUG-EFFLUX TRANSPORTER 1 REGULATOR BMRR | | Authors: | Zheleznova, E.E, Markham, P.N, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter.
Cell(Cambridge,Mass.), 96, 1999
|
|
1RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF UNLIGANDED HUMAN RIBONUCLEASE 4 | | Descriptor: | PROTEIN (RIBONUCLEASE 4) | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 1998-10-29 | | Release date: | 1999-10-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
3D6Y
 
 | |
2VXN
 
 | |
2X16
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2009-12-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1T
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3D6Z
 
 | |
1QQB
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*TP*CP*GP*AP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
2X1R
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-(PROPYLSULFONYL)PROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X2G
 
 | | CRYSTALLOGRAPHIC BINDING STUDIES WITH AN ENGINEERED MONOMERIC VARIANT OF TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1U
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1QP4
 
 | | PURINE REPRESSOR-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*TP*CP*GP*AP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
2X1S
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1DLV
 
 | |
1QP0
 
 | | PURINE REPRESSOR-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*CP*CP*GP*GP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
1QDS
 
 | | SUPERSTABLE E65Q MUTANT OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Lambeir, A.M, Backmann, J, Ruiz-Sanz, J, Filimonov, V, Nielsen, J.E, Vriend, G, Kursula, I, Norledge, B.V, Wierenga, R.K. | | Deposit date: | 1999-07-10 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ionization of a buried glutamic acid is thermodynamically linked to the stability of Leishmania mexicana triose phosphate isomerase.
Eur.J.Biochem., 267, 2000
|
|
1QP7
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*CP*CP*GP*GP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
