2X58
 
 | | The crystal structure of MFE1 liganded with CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, GLYCEROL, ... | | Authors: | Kasaragod, P, Venkatesan, R, Kiema, T.R, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Liganded Rat Peroxisomal Multifunctional Enzyme Type 1: A Flexible Molecule with Two Interconnected Active Sites
J.Biol.Chem., 285, 2010
|
|
3GIB
 
 | |
4E9J
 
 | |
1HAC
 
 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4EC5
 
 | |
2WL6
 
 | |
4NEM
 
 | | Small molecular fragment bound to crystal contact interface of Interleukin-2 | | Descriptor: | 5-[(2,3-dichlorophenoxy)methyl]furan-2-carboxylic acid, Interleukin-2 | | Authors: | Jehle, S, Brenke, R, Vajda, S, Allen, K.N, Kozakov, D. | | Deposit date: | 2013-10-29 | | Release date: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Small molecular fragments bound to binding energy hot-spot in crystal contact interface of Interleukin-2
To be Published
|
|
2V2C
 
 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed-Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VEI
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEM
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | (3-bromo-2-oxo-propoxy)phosphonic acid, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2V2D
 
 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed- Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2XO1
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to N6-methyladenine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine riboswitch, ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2VTZ
 
 | | Biosynthetic thiolase from Z. ramigera. Complex of the C89A mutant with coenzyme A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
3GZE
 
 | |
2XO0
 
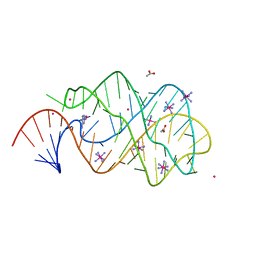 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to 24-diamino-1,3,5- triazine identified by virtual screening | | Descriptor: | 1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2Y63
 
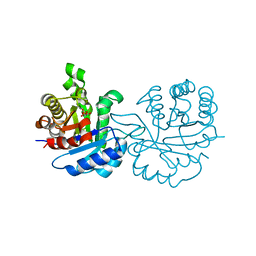 | | Crystal structure of Leishmanial E65Q-TIM complexed with Bromohydroxyacetone phosphate | | Descriptor: | (3-bromo-2-oxo-propoxy)phosphonic acid, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Venkatesan, R, Alahuhta, M, Pihko, P.M, Wierenga, R.K. | | Deposit date: | 2011-01-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High Resolution Crystal Structures of Triosephosphate Isomerase Complexed with its Suicide Inhibitors: The Conformational Flexibility of the Catalytic Glutamate in its Closed, Liganded Active Site.
Protein Sci., 20, 2011
|
|
2VEL
 
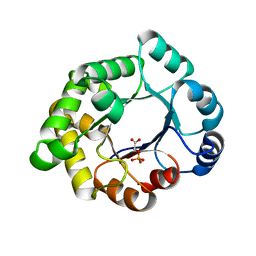 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties. | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2V5F
 
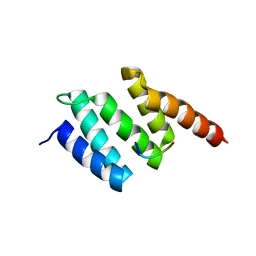 | | Crystal structure of wild type peptide-binding domain of human type I collagen prolyl 4-hydroxylase. | | Descriptor: | HEXA-HISTIDINE PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Pekkala, M, Hieta, R, Kivirikko, K, Myllyharju, J, Wierenga, R. | | Deposit date: | 2008-10-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Wild Type Peptide-Binding Domain of Human Type I Collagen Prolyl 4- Hydroxylase.
To be Published
|
|
2XNW
 
 | | XPT-PBUX C74U RIBOSWITCH FROM B. SUBTILIS BOUND TO A TRIAZOLO- TRIAZOLE-DIAMINE LIGAND IDENTIFIED BY VIRTUAL SCREENING | | Descriptor: | 3,6-diamino-1,5-dihydro[1,2,4]triazolo[4,3-b][1,2,4]triazol-4-ium, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
3GN2
 
 | | Structure of Pteridine Reductase 1 (PTR1) from TRYPANOSOMA BRUCEI in ternary complex with cofactor (NADP+) and inhibitor (DDD00066730) | | Descriptor: | 1-(3,4-dichlorobenzyl)-1H-benzimidazol-2-amine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Brenk, R, Hunter, W.N. | | Deposit date: | 2009-03-16 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening.
J.Med.Chem., 52, 2009
|
|
2XNZ
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to acetoguanamine identified by virtual screening | | Descriptor: | 6-METHYL-1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2Y5A
 
 | | Cytochrome c peroxidase (CCP) W191G bound to 3-aminopyridine | | Descriptor: | 3-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Cappel, D, Wahlstrom, R, Brenk, R, Sotriffer, C.A. | | Deposit date: | 2011-01-12 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the Dynamic Nature of Water Molecules and Their Influences on Ligand Binding in a Model Binding Site.
J.Chem.Inf.Model, 51, 2011
|
|
3GN1
 
 | | Structure of Pteridine Reductase 1 (PTR1) from TRYPANOSOMA BRUCEI in ternary complex with cofactor (NADP+) and inhibitor (DDD00067116) | | Descriptor: | 1H-benzimidazol-2-amine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Brenk, R, Hunter, W.N. | | Deposit date: | 2009-03-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-09-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening.
J.Med.Chem., 52, 2009
|
|
4BI9
 
 | | Crystal structure of wild-type SCP2 thiolase from Trypanosoma brucei. | | Descriptor: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
1K39
 
 | | The structure of yeast delta3-delta2-enoyl-COA isomerase complexed with octanoyl-COA | | Descriptor: | OCTANOYL-COENZYME A, PHOSPHATE ION, d3,d2-enoyl CoA isomerase ECI1 | | Authors: | Mursula, A.M, Geerlof, A, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: |
|
|
