1CK3
 
 | | N276D MUTANT OF ESCHERICHIA COLI TEM-1 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE | | Authors: | Swaren, P, Maveyraud, L, Samama, J.P. | | Deposit date: | 1999-04-27 | | Release date: | 1999-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray structure of the Asn276Asp variant of the Escherichia coli TEM-1 beta-lactamase: direct observation of electrostatic modulation in resistance to inactivation by clavulanic acid.
Biochemistry, 38, 1999
|
|
1WA8
 
 | | Solution Structure of the CFP-10.ESAT-6 Complex. Major Virulence Determinants of Pathogenic Mycobacteria | | Descriptor: | 6 KDA EARLY SECRETORY ANTIGENIC TARGET (ESAT-6), ESAT-6 LIKE PROTEIN ESXB | | Authors: | Renshaw, P.S, Lightbody, K.L, Veverka, V, Muskett, F.W, Kelly, G, Frenkiel, T.A, Gordon, S.V, Hewinson, R.G, Burke, B, Norman, J, Williamson, R.A, Carr, M.D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-25 | | Release date: | 2005-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of the Complex Formed by the Tuberculosis Virulence Factors Cfp-10 and Esat-6
Embo J., 24, 2005
|
|
2XYF
 
 | | HIV-1 Inhibitors with a Tertiary-Alcohol-containing Transition-State Mimic and various P2 and P1 prime Substituents | | Descriptor: | METHYL N-[(2S)-1-[2-[(4R)-5-[[(2S)-3,3-DIMETHYL-1-METHYLAMINO-1-OXO-BUTAN-2-YL]AMINO]-4-HYDROXY-5-OXO-4-(PHENYLMETHYL)PENTYL]-2-[(4-THIOPHEN-3-YLPHENYL)METHYL]HYDRAZINYL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, PROTEASE | | Authors: | Ohrngren, P, Wu, X, Persson, M, Ekegren, J.K, Wallberg, H, Rosenquist, A, Samuelsson, B, Unge, T, Larhed, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 Protease Inhibitors with a Tertiary Alcohol Containing Transition-State Mimic and Various P2 and P1' Substituents
Med.Chem.Commun., 2, 2011
|
|
2XYE
 
 | | HIV-1 Inhibitors with a Tertiary-Alcohol-containing Transition-State Mimic and various P2 and P1 prime Substituents | | Descriptor: | METHYL N-[(2S)-1-[2-[(4R)-5-[[(2S)-3,3-DIMETHYL-1-METHYLAMINO-1-OXO-BUTAN-2-YL]AMINO]-4-HYDROXY-5-OXO-4-(PHENYLMETHYL)PENTYL]-2-[(4-PHENYLPHENYL)METHYL]HYDRAZINYL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, PROTEASE | | Authors: | Ohrngren, P, Wu, X, Persson, M, Ekegren, J.K, Wallberg, H, Rosenquist, A, Samuelsson, B, Unge, T, Larhed, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors with a Tertiary Alcohol Containing Transition-State Mimic and Various P2 and P1' Substituents
Med.Chem.Commun., 2, 2011
|
|
7U1N
 
 | | Crystal structure of the Anopheles darlingi AD-118 long form D7 salivary protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-ETHOXYETHANOL, ... | | Authors: | Alvarenga, P.H, Gittis, A.G, Garboczi, D.N, Andersen, J.F. | | Deposit date: | 2022-02-21 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional aspects of evolution in a cluster of salivary protein genes from mosquitoes.
Insect Biochem.Mol.Biol., 146, 2022
|
|
2NDH
 
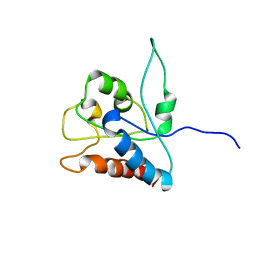 | | NMR solution structure of MAL/TIRAP TIR domain (C116A) | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Lavrencic, P, Mobli, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3RDT
 
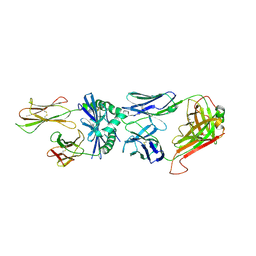 | | Crystal Structure of 809.B5 TCR complexed with MHC Class II I-Ab/3k peptide | | Descriptor: | 3K peptide, linker and MHC H-2 class II I-Ab beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Trenh, P, Huseby, E.S, Stern, L.J. | | Deposit date: | 2011-04-01 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands.
Immunity, 35, 2011
|
|
1BUE
 
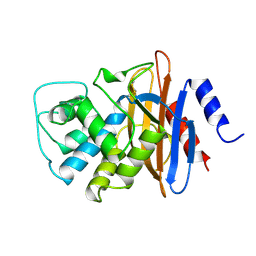 | | NMC-A CARBAPENEMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | PROTEIN (IMIPENEM-HYDROLYSING BETA-LACTAMASE) | | Authors: | Swaren, P, Maveyraud, L, Cabantous, S, Pedelacq, J.D, Mourey, L, Frere, J.M, Samama, J.P. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray analysis of the NMC-A beta-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity.
J.Biol.Chem., 273, 1998
|
|
2NCG
 
 | |
4P5T
 
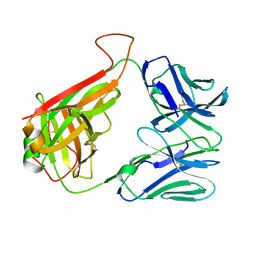 | | 14.C6 TCR complexed with MHC class II I-Ab/3K peptide | | Descriptor: | H-2 class II histocompatibility antigen, A-B alpha chain, Human nkt tcr beta chain, ... | | Authors: | Trenh, P, Stadinski, B, Huseby, E.S, Stern, L.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
4OV5
 
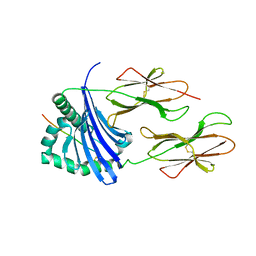 | | Structure of HLA-DR1 with a bound peptide with non-optimal alanine in the P1 pocket | | Descriptor: | HLA class I histocompatibility antigen, A-2 alpha chain, HLA class II histocompatibility antigen, ... | | Authors: | Trenh, P, Yin, L, Stern, L.J. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Susceptibility to HLA-DM Protein Is Determined by a Dynamic Conformation of Major Histocompatibility Complex Class II Molecule Bound with Peptide.
J.Biol.Chem., 289, 2014
|
|
3ZRT
 
 | | Crystal structure of human PSD-95 PDZ1-2 | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Sorensen, P.L, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2011-06-19 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | A High-Affinity, Dimeric Inhibitor of Psd-95 Bivalently Interacts with Pdz1-2 and Protects Against Ischemic Brain Damage.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7WVK
 
 | | Crystal structure of human WDR5 in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,5-bis(chloranyl)phenyl]sulfonylbenzimidazole, GLYCEROL, ... | | Authors: | Han, Q.L, Zhang, X.L, Wang, L, Ren, P.X, Cao, Y, Li, K, Bai, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery, evaluation and mechanism study of WDR5-targeted small molecular inhibitors for neuroblastoma.
Acta Pharmacol.Sin., 44, 2023
|
|
6B0V
 
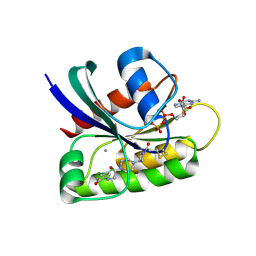 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0Y
 
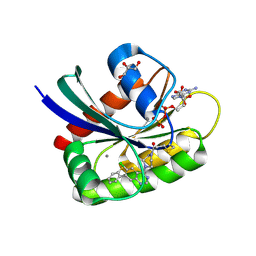 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
5PB7
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09440a | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indazol-5-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PBG
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 1) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PBT
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 14) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PCA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 31) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PCK
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 41) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PCX
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 54) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PBK
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 5) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDE
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 70) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
