7UR5
 
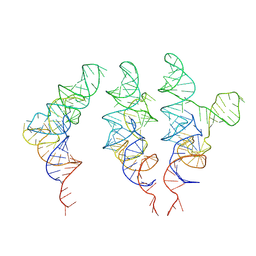 | | allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome | | Descriptor: | allo-tRNAUTu1 | | Authors: | Zhang, J, Krahn, N, Prabhakar, A, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7URM
 
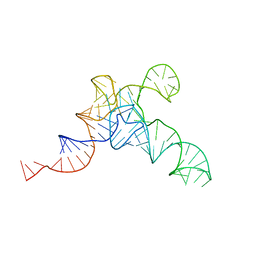 | | allo-tRNAUTu1A in the P site of the E. coli ribosome | | Descriptor: | allo-tRNAUTU1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
5A7M
 
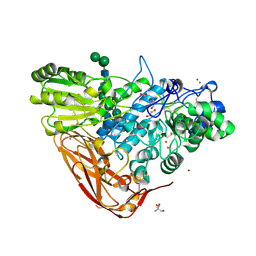 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
7RK6
 
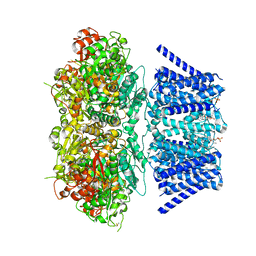 | | Aplysia Slo1 with Barium | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BARIUM ION, BK channel, ... | | Authors: | Zhu, J, Srivastava, S, Cachau, R, Holmgren, M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | CryoEM structure of Aplysia Slo1 with 0 mM Ba2+ at 2.91 A
To Be Published
|
|
7RJT
 
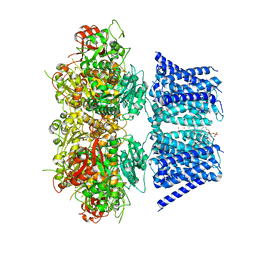 | | Aplysia Slo1 with Barium | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BARIUM ION, BK channel, ... | | Authors: | Zhu, J, Srivastava, S, Cachau, R, Holmgren, M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | CryoEM structure of Aplysia Slo1 with 10 mM Ba2+ at 2.93 A
To Be Published
|
|
5MDU
 
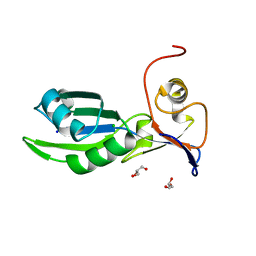 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe. | | Descriptor: | CHLORIDE ION, GLYCEROL, Rpb7-binding protein seb1, ... | | Authors: | Wittmann, S, Renner, M, El Omari, K, Adams, O, Vasiljeva, L, Grimes, J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA.
Nat Commun, 8, 2017
|
|
5MDT
 
 | |
7Z79
 
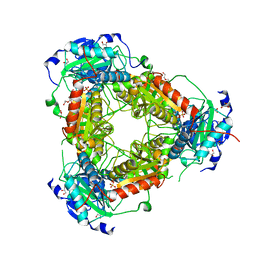 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
8B3Q
 
 | |
8B3P
 
 | |
8B3O
 
 | |
7AY2
 
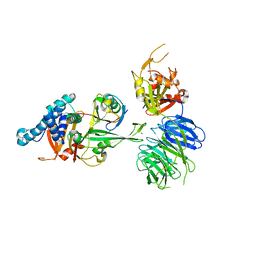 | | Crystal structure of truncated USP1-UAF1 reacted with ubiquitin-prg | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 1, WD repeat-containing protein 48, ... | | Authors: | Arkinson, C, Rennie, M.L, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7S8V
 
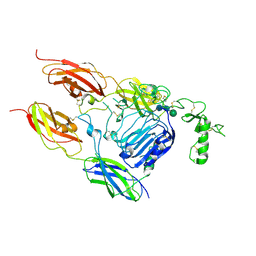 | |
7S0Q
 
 | |
4IRA
 
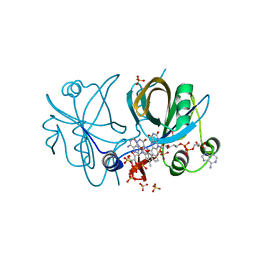 | | CobR in complex with FAD | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Lawrence, A.D, Scott, A.F, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biophysical characterisation and structure-function analysis of Brucella melitensis CobR: Protein-flavin interactions determine function and stability
To be Published
|
|
6Y0J
 
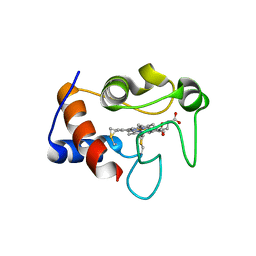 | |
5NNS
 
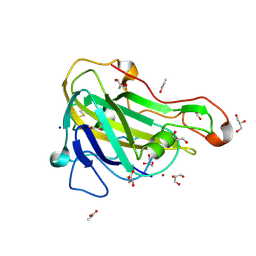 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
7OC8
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with pNPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
7NYT
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with lactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-03-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
8PDP
 
 | | 10-mer ring of HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDM
 
 | | 11-mer ring of human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
8OOD
 
 | |
7ZF1
 
 | | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Lemonidis, K, Rennie, M.L, Arkinson, C, Streetley, J, Clarke, M, Chaugule, V.K, Walden, H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Embo J., 42, 2023
|
|
8OO5
 
 | |
