7M27
 
 | |
7M2C
 
 | |
7M2B
 
 | |
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
7RC7
 
 | | Solution NMR Structure of [Ala19]Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Conibear, A.C, Rosengren, K.J. | | Deposit date: | 2021-07-07 | | Release date: | 2021-08-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A conserved beta-bulge glycine residue facilitates folding and increases stability of the mouse alpha-defensin cryptdin-4
Peptide Science, 114, 2022
|
|
6ENT
 
 | | Structure of the rat RKIP variant delta143-146 | | Descriptor: | CHLORIDE ION, Phosphatidylethanolamine-binding protein 1, ZINC ION | | Authors: | Koelmel, W, Hirschbeck, M, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ENS
 
 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2ACQ
 
 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
2ACS
 
 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
2ACR
 
 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CACODYLATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
4RVY
 
 | | Serial Time resolved crystallography of Photosystem II using a femtosecond X-ray laser. The S state after two flashes (S3) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.-H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-11-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
2ATG
 
 | | NMR structure of Retrocyclin-2 in SDS | | Descriptor: | Retrocyclin-2 | | Authors: | Daly, N.L, Chen, Y.K, Rosengren, K.J, Marx, U.C, Phillips, M.L, Waring, A.J, Wang, W, Lehrer, R.I, Craik, D.J. | | Deposit date: | 2005-08-24 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin
Biochemistry, 46, 2007
|
|
2Q3N
 
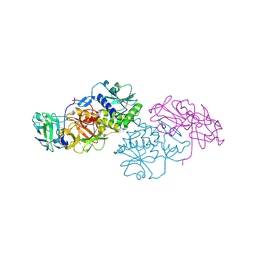 | | Agglutinin from Abrus Precatorius (APA-I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 A chain, Agglutinin-1 B chain | | Authors: | Bagaria, A, Surendranath, K, Ramagopal, U.A, Ramakumar, S, Karande, A.A. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Function Analysis and Insights into the Reduced Toxicity of Abrus precatorius Agglutinin I in Relation to Abrin.
J.Biol.Chem., 281, 2006
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4X
 
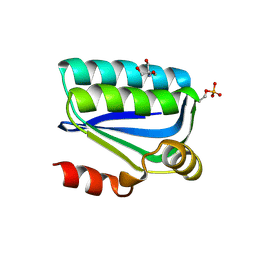 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
4XCM
 
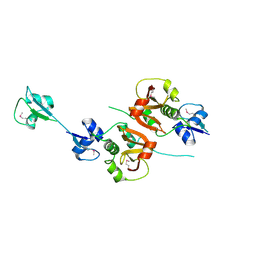 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X8D
 
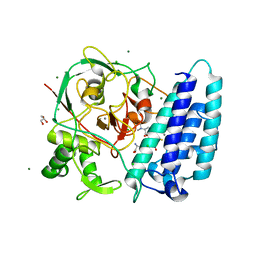 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB in complex with N,N-dimethyl-histidine and gamma-glutamyl-cysteine | | Descriptor: | CALCIUM ION, CHLORIDE ION, GAMMA-GLUTAMYLCYSTEINE, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4X8B
 
 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB, apo form | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4X8E
 
 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB in complex with N,N,N-trimethyl-histidine | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OPB
 
 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
