3UVF
 
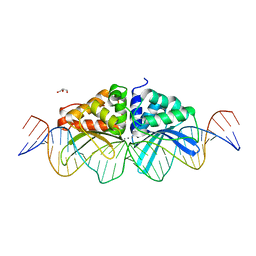 | | Expanding LAGALIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Intron-encoded DNA endonuclease I-HjeMI, ... | | Authors: | Jacoby, K, Metzger, M, Shen, B, Jarjour, J, Stoddard, B, Scharenberg, A. | | Deposit date: | 2011-11-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space.
Nucleic Acids Res., 40, 2012
|
|
2XII
 
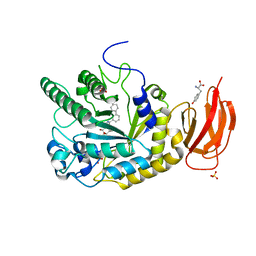 | | CRYSTAL STRUCTURE OF AN ALPHA-L-FUCOSIDASE GH29 FROM BACTEROIDES THETAIOTAOMICRON IN COMPLEX WITH AN EXTENDED 9-FLUORENONE IMINOSUGAR INHIBITOR | | Descriptor: | 9-oxo-N-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyl-piperidin-2-yl]methyl]fluorene-1-carboxamide, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Popat, S.D, Lin, C.H, Davies, G.J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Thermodynamic Analyses of Alpha-L-Fucosidase Inhibitors.
Chembiochem, 11, 2010
|
|
1R70
 
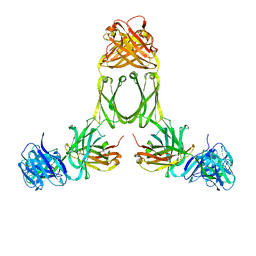 | | Model of human IgA2 determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Human IgA2(m1) Heavy Chain, Human IgA2(m1) Light Chain | | Authors: | Furtado, P.B, Whitty, P.W, Robertson, A, Eaton, J.T, Almogren, A, Kerr, M.A, Woof, J.M, Perkins, S.J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | Solution Structure Determination of Monomeric Human IgA2 by X-ray and Neutron Scattering, Analytical Ultracentrifugation and Constrained Modelling: A Comparison with Monomeric Human IgA1.
J.Mol.Biol., 338, 2004
|
|
2NNR
 
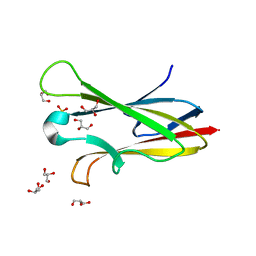 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
1YKA
 
 | | Solution structure of Grx4, a monothiol glutaredoxin from E. coli. | | Descriptor: | monothiol glutaredoxin ydhD | | Authors: | Fladvad, M, Bellanda, M, Fernandes, A.P, Andresen, C, Mammi, S, Holmgren, A, Vlamis-Gardikas, A, Sunnerhagen, M. | | Deposit date: | 2005-01-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mapping of functionalities in the solution structure of reduced Grx4, a monothiol glutaredoxin from Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1Q33
 
 | | Crystal structure of human ADP-ribose pyrophosphatase NUDT9 | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, beta-D-glucopyranose | | Authors: | Shen, B.W, Perraud, A.L, Scharenberg, A, Stoddard, B.L. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure and Mutational Analysis of Human NUDT9
J.Mol.Biol., 332, 2003
|
|
2B4Z
 
 | | Crystal structure of cytochrome C from bovine heart at 1.5 A resolution. | | Descriptor: | Cytochrome c, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mirkin, N, Jakoncic, J, Stojanoff, V, Moreno, A. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor.
Proteins, 70, 2008
|
|
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
5A23
 
 | | SdsA sulfatase triclinic form | | Descriptor: | SDS HYDROLASE SDSA1, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncik, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-05-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
1L9L
 
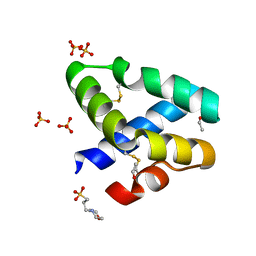 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1UY4
 
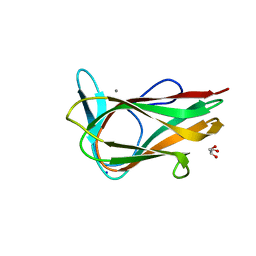 | |
1UY2
 
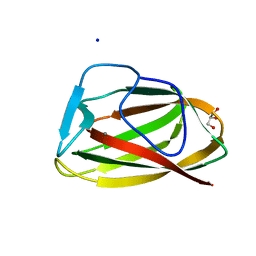 | |
1UY3
 
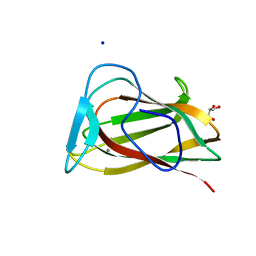 | |
1UY1
 
 | |
2HVC
 
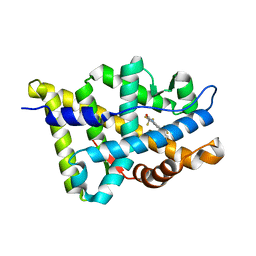 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
1U4R
 
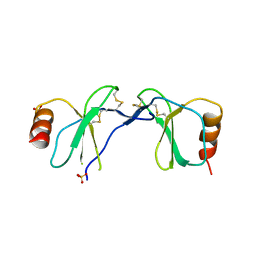 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
2P3X
 
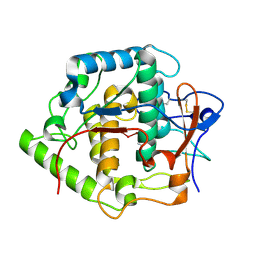 | | Crystal structure of Grenache (Vitis vinifera) Polyphenol Oxidase | | Descriptor: | CU-O-CU LINKAGE, Polyphenol oxidase, chloroplast | | Authors: | Reyes Grajeda, J.P, Virador, V.M, Blanco-Labra, A, Mendiola-Olaya, E, Smith, G.M, Moreno, A, Whitaker, J.R. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, purification, and crystal structure of Grenache (Vitis vinifera) polyphenol oxidase.
J.Agric.Food Chem., 58, 2010
|
|
1GZ2
 
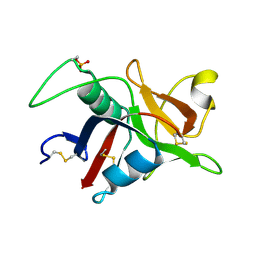 | |
3BEW
 
 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
3ZM8
 
 | | Crystal structure of Podospora anserina GH26-CBM35 beta-(1,4)- mannanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GH26 ENDO-BETA-1,4-MANNANASE, ... | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
1H6V
 
 | | Mammalian thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | Authors: | Sandalova, T, Zhong, L, Lindqvist, Y, Holmgren, A, Schneider, G. | | Deposit date: | 2001-06-27 | | Release date: | 2001-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of a Mammalian Thioredoxin Reductase: Implication for Mechanism and Evolution of a Selenocysteine Dependent Enzyme
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2Q1Q
 
 | | Carbonic anhydrase inhibitors. Interaction of the antiepileptic drug sulthiame with twelve mammalian isoforms: kinetic and X-Ray crystallographic studies | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, SULTHIAME, ... | | Authors: | Temperini, C, Innocenti, A, Mastrolorenzo, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2007-05-25 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of the antiepileptic drug sulthiame with twelve mammalian isoforms: Kinetic and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
7P82
 
 | | Crystal structure of apo form L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P84
 
 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
