5DZA
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA C terminal domain (WT) | | Descriptor: | 1,2-ETHANEDIOL, BspA, DI(HYDROXYETHYL)ETHER | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
5DZ8
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA variable (V) domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BspA (BspA_V), ... | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
5DZ9
 
 | |
5V8I
 
 | | Thermus thermophilus 70S ribosome lacking ribosomal protein uS17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Gregory, S.T, Jogl, G. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural robustness of the ribosome inferred from X-ray crystal structures of the 30S ribosomal subunit and the 70S ribosome lacking ribosomal protein uS17
To be published
|
|
2M5F
 
 | |
9C0O
 
 | |
2LCY
 
 | |
2LCZ
 
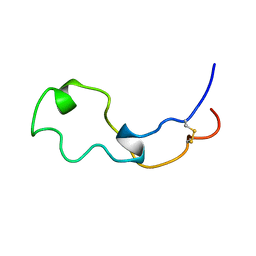 | |
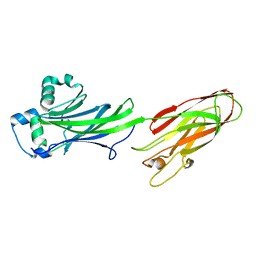
4Z23
 
 | | BspA_C_WT | | Descriptor: | 1,2-ETHANEDIOL, Cell wall surface anchor protein | | Authors: | Race, P, Rego, S. | | Deposit date: | 2015-03-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BspA_C_WT
To Be Published
|
|
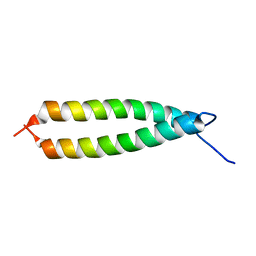
4Z1P
 
 | | BspA_C_mut | | Descriptor: | Cell wall surface anchor protein | | Authors: | Race, P, Rego, S. | | Deposit date: | 2015-03-27 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BspA_C_mut
To Be Published
|
|
1IHQ
 
 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | Descriptor: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | Authors: | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
3CJR
 
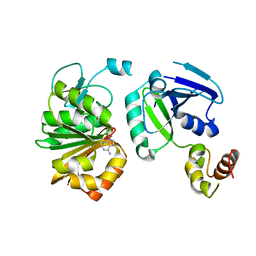 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with ribosomal protein L11 (K39A) and inhibitor Sinefungin. | | Descriptor: | 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase, SINEFUNGIN | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
3CJS
 
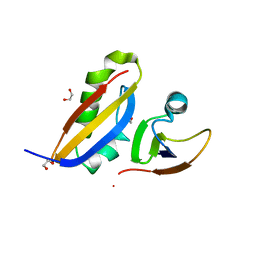 | | Minimal Recognition Complex between PrmA and Ribosomal Protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
1TMZ
 
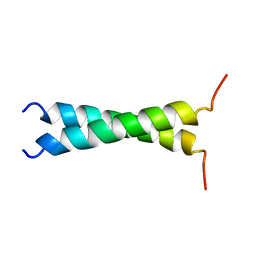 | | TMZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF ALPHA TROPOMYOSIN, NMR, 15 STRUCTURES | | Descriptor: | TMZIP | | Authors: | Greenfield, N.J, Montelione, G.T, Hitchcock-Degregori, S.E, Farid, R.S. | | Deposit date: | 1998-04-20 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies.
Biochemistry, 37, 1998
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
1MV4
 
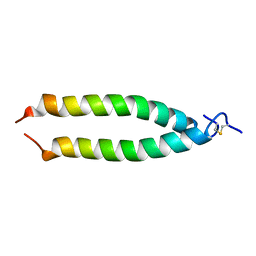 | | TM9A251-284: A Peptide Model of the C-Terminus of a Rat Striated Alpha Tropomyosin | | Descriptor: | Tropomyosin 1 alpha chain | | Authors: | Greenfield, N.J, Swapna, G.V.T, Huang, Y, Palm, T, Graboski, S, Montelione, G.T, Hitchcock-Degregori, S.E. | | Deposit date: | 2002-09-24 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Carboxyl Terminus of Striated alpha-Tropomyosin in Solution Reveals an Unusual Parallel Arrangement of Interacting alpha-Helices
Biochemistry, 42, 2003
|
|
1IC2
 
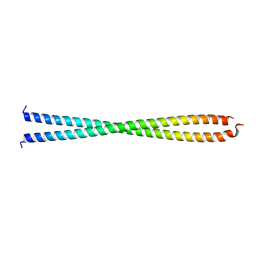 | | DECIPHERING THE DESIGN OF THE TROPOMYOSIN MOLECULE | | Descriptor: | TROPOMYOSIN ALPHA CHAIN, SKELETAL MUSCLE | | Authors: | Brown, J.H, Kim, K.-H, Jun, G, Greenfield, N.J, Dominguez, R, Volkmann, N, Hitchcock-DeGregori, S.E, Cohen, C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the design of the tropomyosin molecule
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7JNH
 
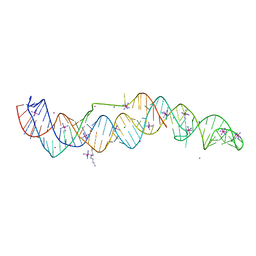 | | Crystal structure of a double-ENE RNA stability element in complex with a 28-mer poly(A) RNA | | Descriptor: | 28-mer poly(A) RNA, COBALT HEXAMMINE(III), Core double ENE RNA (Xtal construct) from Oryza sativa transposon,Core double ENE RNA (Xtal construct) from Oryza sativa transposon, ... | | Authors: | Torabi, S.F, Vaidya, A.T, Tycowski, K.T, DeGregorio, S.J, Wang, J, Shu, M.D, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction.
Science, 371, 2021
|
|
2K8X
 
 | |
3P22
 
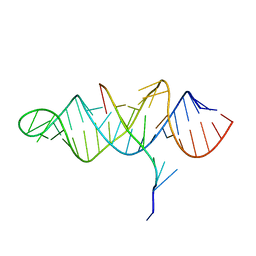 | | Crystal structure of the ENE, a viral RNA stability element, in complex with A9 RNA | | Descriptor: | Core ENE hairpin from KSHV PAN RNA, oligo(A)9 RNA | | Authors: | Mitton-Fry, R.M, DeGregorio, S.J, Wang, J, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2010-10-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Poly(A) tail recognition by a viral RNA element through assembly of a triple helix.
Science, 330, 2010
|
|
3CJT
 
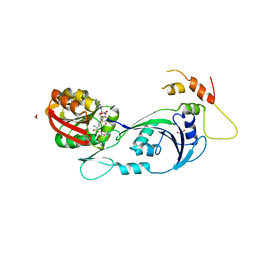 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, CHLORIDE ION, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
6XHX
 
 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with erythromycin and protein Y (YfiA) at 2.55A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XHV
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XHW
 
 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XHY
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with telithromycin, mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
