3TX8
 
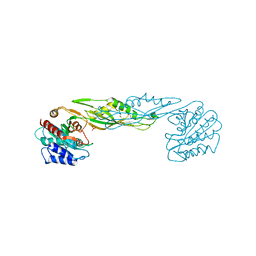 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1B3K
 
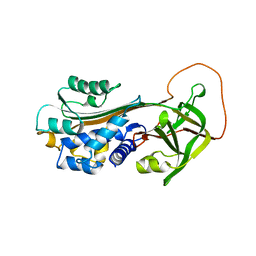 | | Plasminogen activator inhibitor-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Sharp, A.M, Stein, P.E, Pannu, N.S, Read, R.J. | | Deposit date: | 1998-12-11 | | Release date: | 1999-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion.
Structure Fold.Des., 7, 1999
|
|
1CZG
 
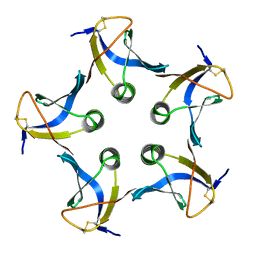 | |
1CZW
 
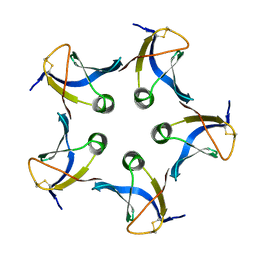 | |
2R82
 
 | | Pyruvate phosphate dikinase (PPDK) triple mutant R219E/E271R/S262D adapts a second conformational state | | Descriptor: | Pyruvate, phosphate dikinase, SULFATE ION | | Authors: | Lim, K, Read, R.J, Chen, C.C, Herzberg, O. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Swiveling domain mechanism in pyruvate phosphate dikinase.
Biochemistry, 46, 2007
|
|
2RIV
 
 | | Crystal structure of the reactive loop cleaved human Thyroxine Binding Globulin | | Descriptor: | GLYCEROL, SULFATE ION, Thyroxine-binding globulin | | Authors: | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | Deposit date: | 2007-10-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
2RIW
 
 | | The Reactive loop cleaved human Thyroxine Binding Globulin complexed with thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, Thyroxine-binding globulin | | Authors: | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | Deposit date: | 2007-10-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
2VDY
 
 | | Crystal structure of the reactive loop cleaved Corticosteroid Binding Globulin complexed with Cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, CORTICOSTEROID-BINDING GLOBULIN | | Authors: | Zhou, A, Wei, Z, Read, R.J. | | Deposit date: | 2007-10-13 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The S-to-R Transition of Corticosteroid-Binding Globulin and the Mechanism of Hormone Release.
J.Mol.Biol., 380, 2008
|
|
2VDX
 
 | |
2WXW
 
 | |
2WY1
 
 | |
2WXX
 
 | |
2WY0
 
 | | Crystal structure of mouse angiotensinogen in the oxidised form with space group P6122 | | Descriptor: | 1,2-ETHANEDIOL, ANGIOTENSINOGEN, SODIUM ION | | Authors: | Zhou, A, Wei, Z, Carrell, R.W, Read, R.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A Redox Switch in Angiotensinogen Modulates Angiotensin Release.
Nature, 468, 2010
|
|
2WXZ
 
 | |
3ZR5
 
 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | Deposit date: | 2011-06-14 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZR6
 
 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | Deposit date: | 2011-06-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3MIW
 
 | | Crystal Structure of Rotavirus NSP4 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural glycoprotein 4 | | Authors: | Chacko, A.R, Read, R.J, Dodson, E.J, Rao, D.C, Suguna, K. | | Deposit date: | 2010-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new pentameric structure of rotavirus NSP4 revealed by molecular replacement.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4N3E
 
 | | Crystal structure of Hyp-1, a St John's wort PR-10 protein, in complex with 8-anilino-1-naphthalene sulfonate (ANS) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Mccoy, A.J, Read, R.J, Jaskolski, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Likelihood-based molecular-replacement solution for a highly pathological crystal with tetartohedral twinning and sevenfold translational noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1QLP
 
 | | 2.0 ANGSTROM STRUCTURE OF INTACT ALPHA-1-ANTITRYPSIN: A CANONICAL TEMPLATE FOR ACTIVE SERPINS | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Elliott, P.R, Pei, X.Y, Dafforn, T, Read, R.J, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1999-09-10 | | Release date: | 1999-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease.
Protein Sci., 9, 2000
|
|
