8BC1
 
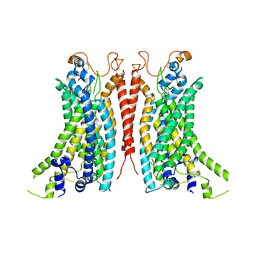 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
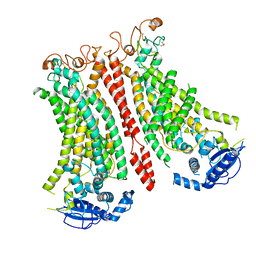 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8Q
 
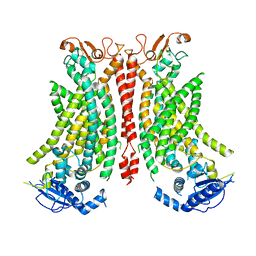 | | Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8YQ9
 
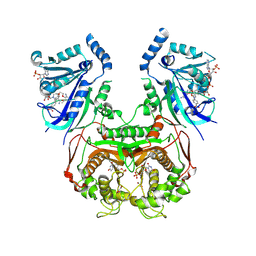 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB6, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxypropoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
8ONT
 
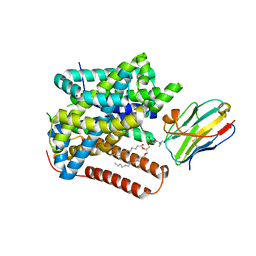 | | Structure of Setaria italica NRAT in complex with a nanobody | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, NRAMP related aluminium transporter, Nanobody1 | | Authors: | Ramanadane, K, Liziczai, M, Markovic, D, Straub, M.S, Rosalen, G.T, Udovcic, A, Dutzler, R, Manatschal, C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural and functional properties of a plant NRAMP-related aluminum transporter.
Elife, 12, 2023
|
|
4QS5
 
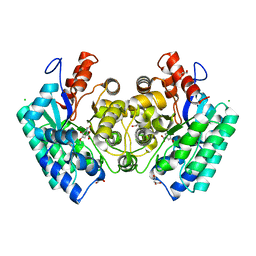 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND MANGANESE AND 3-methoxy-4-hydroxy-5-nitrobenzoic acid, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 5-CARBOXYVANILLATE Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QRO
 
 | | CRYSTAL STRUCTURE of DIHYDROXYBENZOIC ACID DECARBBOXYLASE BPRO_2061 (TARGET EFI-500288) FROM POLAROMONAS SP. JS666 WITH BOUND MANGANESE AND AN INHIBITOR, 2-NITRORESORCINOL | | Descriptor: | 2-nitrobenzene-1,3-diol, ACETATE ION, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Dihydroxybenzoate Decarboxylase from Frompolaromonas Sp WITH BOUND MANGANESE AND 2-NITRORESORCINOL
To be Published
|
|
1TOS
 
 | | TORPEDO CALIFORNICA ACHR RECEPTOR [ALA76] ANALOGUE COMPLEXED WITH THE ANTI-ACETYLCHOLINE MAB6 MONOCLONAL ANTIBODY | | Descriptor: | ACETYLCHOLINE RECEPTOR [ALA76] MIR ANALOGUE | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
2RO4
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO3
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator Abh | | Descriptor: | Putative transition state regulator abh | | Authors: | Sullivan, D.M, Bobay, B.G, Douglas, K.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
1ACF
 
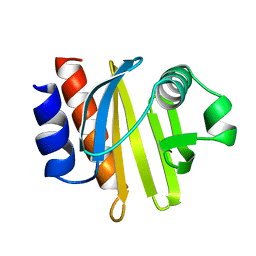 | | ACANTHAMOEBA CASTELLANII PROFILIN IB | | Descriptor: | PROFILIN I | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-07-29 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1KFX
 
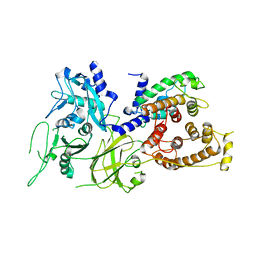 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8P5I
 
 | | Kinase domain of mutant human ULK1 in complex with compound XMD-17-51 | | Descriptor: | 5,11-dimethyl-2-[(1-piperidin-4-ylpyrazol-4-yl)amino]pyrimido[4,5-b][1,4]benzodiazepin-6-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8P5K
 
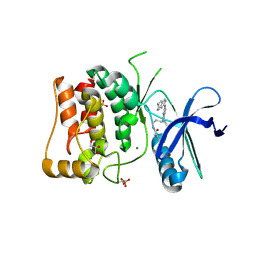 | | Kinase domain of mutant human ULK1 in complex with compound MRT68921 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
1Y2M
 
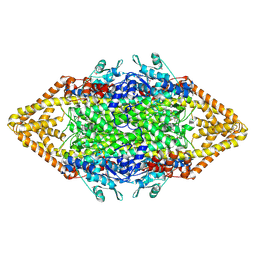 | | Crystal structure of phenylalanine ammonia-lyase from yeast Rhododporidium toruloides | | Descriptor: | Phenylalanine ammonia-lyase | | Authors: | Wang, L, Gamez, A, Sarkissian, C.N, Straub, M, Patch, M.G, Han, G.W, Scriver, C.R, Stevens, R.C. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria.
Mol.Genet.Metab., 86, 2005
|
|
1KFU
 
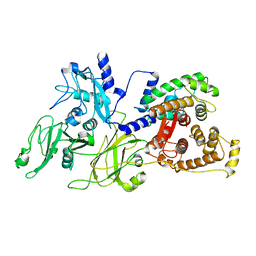 | | Crystal Structure of Human m-Calpain Form II | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KET
 
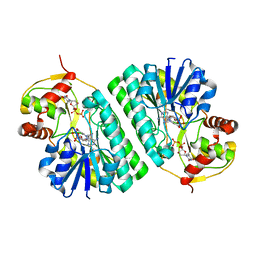 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with thymidine diphosphate bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KEU
 
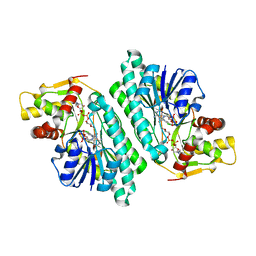 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KEP
 
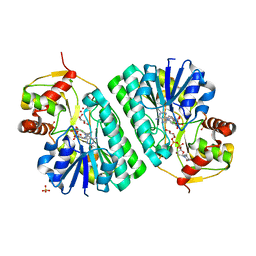 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-xylose bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
3ZTR
 
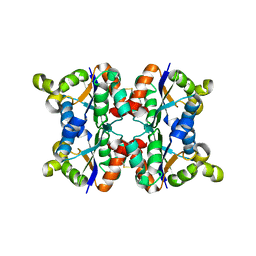 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FIRST STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
1KER
 
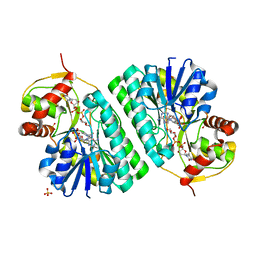 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
3ZTO
 
 | | Orthorhombic crystal form C222 of the Aquifex aeolicus nucleoside diphosphate kinase | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | An Intersubunit Disulfide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus.
Proteins, 80, 2012
|
|
3ZTP
 
 | | Orthorhombic crystal form P21212 of the Aquifex aeolicus nucleoside diphosphate kinase | | Descriptor: | GLYCEROL, NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | An Intersubunit Disulfide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus.
Proteins, 80, 2012
|
|
3ZTQ
 
 | | Hexagonal crystal form P61 of the Aquifex aeolicus nucleoside diphosphate kinase | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
3ZTS
 
 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FINAL STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
