7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7CXM
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
2OBS
 
 | |
2OBT
 
 | |
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
2OBR
 
 | | Crystal Structures of P Domain of Norovirus VA387 | | Descriptor: | Capsid protein | | Authors: | Cao, S, Lou, Z, Jiang, X, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2006-12-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of blood group trisaccharides by norovirus.
J.Virol., 81, 2007
|
|
5GNT
 
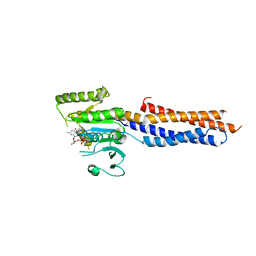 | | BDLP-like folding of Mitofusin 1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Rao, Z, Lou, Z, Hu, J. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | the structure of mini-MFN1 in complex with GDP
To Be Published
|
|
3IWM
 
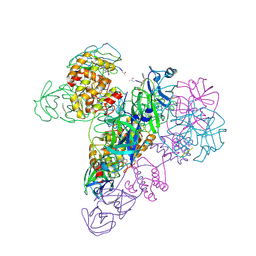 | | The octameric SARS-CoV main protease | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | Deposit date: | 2009-09-02 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
