6L2U
 
 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | Authors: | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | Deposit date: | 2019-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
6LQD
 
 | | Structure of Enterovirus 71 in complex with NLD-22 | | Descriptor: | 1-(2-azanylpyridin-4-yl)-3-[5-[4-(5-methyl-1,2,4-oxadiazol-3-yl)phenoxy]pentyl]imidazolidin-2-one, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Zhang, M, Sun, Y, Wang, X, Guo, Y, Rao, Z. | | Deposit date: | 2020-01-13 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.264 Å) | | Cite: | Design, Synthesis, and Evaluation of Novel Enterovirus 71 Inhibitors as Therapeutic Drug Leads for the Treatment of Human Hand, Foot, and Mouth Disease.
J.Med.Chem., 63, 2020
|
|
1QZ2
 
 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4RVS
 
 | | The native structure of mycobacterial quinone oxidoreductase Rv154c. | | Descriptor: | Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) | | Authors: | Zhou, W.H, Zheng, Q.Q, Song, Y.L, Zhang, W, Shaw, N, Rao, Z. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8464 Å) | | Cite: | Structural views of quinone oxidoreductase from Mycobacterium tuberculosis reveal large conformational changes induced by the co-factor.
Febs J., 282, 2015
|
|
4RM0
 
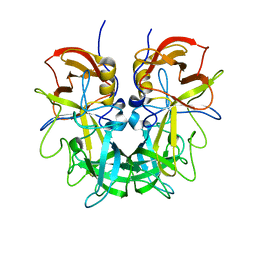 | | Crystal structure of Norovirus OIF P domain in complex with Lewis a trisaccharide | | Descriptor: | Capsid protein, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
4JGZ
 
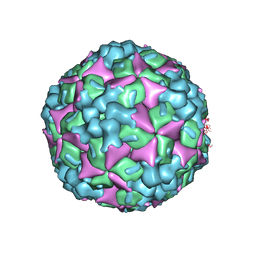 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
3L1N
 
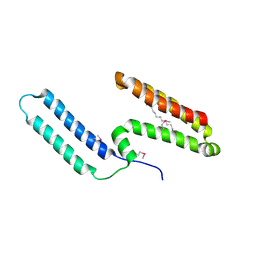 | | Crystal structure of Mp1p ligand binding domain 2 complexd with palmitic acid | | Descriptor: | Cell wall antigen, PALMITIC ACID | | Authors: | Liao, S, Tung, E.T, Zheng, W, Chong, K, Xu, Y, Bartlam, M, Rao, Z, Yuen, K.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mp1p ligand binding domain 2 reveals its function as a fatty acid-binding protein.
J.Biol.Chem., 285, 2010
|
|
1Q1C
 
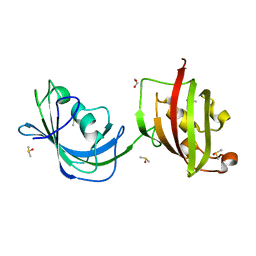 | | Crystal structure of N(1-260) of human FKBP52 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-07-18 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4RVU
 
 | | The native structure of mycobacterial Rv1454c complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) | | Authors: | Zhou, W.H, Zheng, Q.Q, Song, Y.L, Zhang, W, Shaw, N, Rao, Z. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7988 Å) | | Cite: | Structural views of quinone oxidoreductase from Mycobacterium tuberculosis reveal large conformational changes induced by the co-factor.
Febs J., 282, 2015
|
|
4RS4
 
 | |
4RLZ
 
 | | Crystal structure of Norovirus OIF P domain | | Descriptor: | Capsid protein, GLYCEROL | | Authors: | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
3NCO
 
 | | Crystal structure of FnCel5A from F. nodosum Rt17-B1 | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, peptide (ALA)(ASN)(GLU), ... | | Authors: | Zheng, B.S, Yang, W, Wang, Y, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2010-06-05 | | Release date: | 2011-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of FnCel5A from F. nodosum Rt17-B1
To be Published
|
|
1J4I
 
 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
3LXF
 
 | | Crystal Structure of [2Fe-2S] Ferredoxin Arx from Novosphingobium aromaticivorans | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
5GNU
 
 | | the structure of mini-MFN1 apo | | Descriptor: | Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Lou, J. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.113 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
1P5Q
 
 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4JGY
 
 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
7EGQ
 
 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
3PYE
 
 | | Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) in complex with CDPME | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | | Authors: | Shan, S, Chen, X.H, Liu, T, Zhao, H.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis for anti-TB Drug Discovery targeting of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis
To be Published
|
|
5GNR
 
 | | the structure of mini-MFN1 K88A in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Hu, J. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
1J4H
 
 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1ZD7
 
 | | 1.7 Angstrom Crystal Structure Of Post-Splicing Form of a dnaE Intein from Synechocystis Sp. Pcc 6803 | | Descriptor: | DNA polymerase III alpha subunit, SULFATE ION | | Authors: | Sun, P, Ye, S, Ferrandon, S, Evans, T.C, Xu, M.Q, Rao, Z. | | Deposit date: | 2005-04-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing
J.Mol.Biol., 353, 2005
|
|
3RJY
 
 | | Crystal Structure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, alpha-D-glucopyranose | | Authors: | Zheng, B, Yang, W, Zhao, X, Wang, Y, Lou, Z, Rao, Z, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hyperthermophilic Endo-beta-1,4-glucanase: Implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
1UDE
 
 | | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Liu, B, Gao, R, Zhou, W, Bartlam, M, Rao, Z. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii.
Biophys.J., 86, 2004
|
|
