5MY3
 
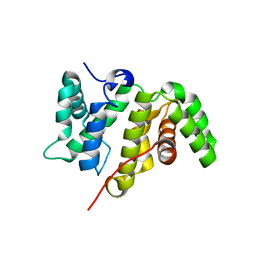 | | Crystal structure of the RhoGAP domain of Rgd1p at 2.19 Angstroms resolution | | Descriptor: | RHO GTPase-activating protein RGD1 | | Authors: | Martinez, D.M, d'Estaintot, B.L, Granier, T, Hugues, M, Odaert, B, Gallois, B, Doignon, F. | | Deposit date: | 2017-01-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural evidence of a phosphoinositide-binding site in the Rgd1-RhoGAP domain.
Biochem. J., 474, 2017
|
|
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Rang, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
6FDL
 
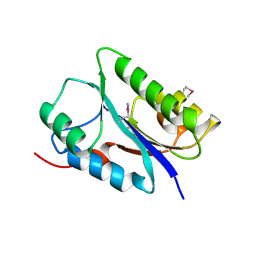 | | Crystal structure of the NYN domain of human MARF1 | | Descriptor: | Meiosis regulator and mRNA stability factor 1 | | Authors: | Jinek, M, Brandmann, T. | | Deposit date: | 2017-12-26 | | Release date: | 2018-11-07 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human MARF1 is an endoribonuclease that interacts with the DCP1:2 decapping complex and degrades target mRNAs.
Nucleic Acids Res., 46, 2018
|
|
5LVS
 
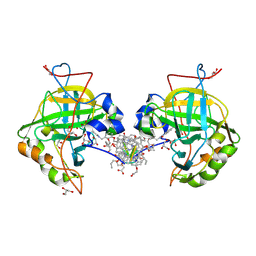 | | Self-assembled protein-aromatic foldamer complexes with 2:3 and 2:2:1 stoichiometries | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, ... | | Authors: | Jewginski, M, LANGLOIS D'ESTAINTOT, B, Granier, T, Huc, Y. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
5NW6
 
 | |
6Q9T
 
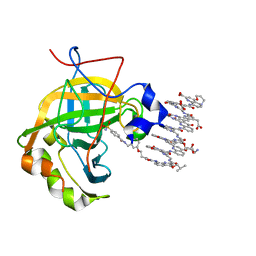 | | Protein-aromatic foldamer complex crystal structure | | Descriptor: | 2-(8-azanyl-2-methanoyl-quinolin-4-yl)ethanoic acid, 6-(aminomethyl)pyridine-2-carboxylic acid, 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, ... | | Authors: | Post, S, Langlois d'Estaintot, B, Fischer, L, Granier, T, Huc, I. | | Deposit date: | 2018-12-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure Elucidation of Helical Aromatic Foldamer-Protein Complexes with Large Contact Surface Areas.
Chemistry, 25, 2019
|
|
5H69
 
 | |
5NVI
 
 | |
5O27
 
 | |
5NSW
 
 | | Xenon for tunnelling analysis of the efflux pump component OprN. | | Descriptor: | Multidrug efflux outer membrane protein OprN, NICKEL (II) ION, PALMITIC ACID, ... | | Authors: | Phan, G, Prange, T, Enguene Ntsogo, Y.V, Garnier, C, Ducruix, A, Broutin, I. | | Deposit date: | 2017-04-27 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
1XY3
 
 | | Urate oxidase from aspergillus flavus complexed with guanine | | Descriptor: | GUANINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-11-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4OP9
 
 | | Urate OXIDASE IN COMPLEX WITH 8-AZAXANTHINE | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Prange, T. | | Deposit date: | 2014-02-05 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional relevance of the internal hydrophobic cavity of urate oxidase.
Febs Lett., 588, 2014
|
|
3P9F
 
 | | Urate oxidase-azaxanthine-azide ternary complex | | Descriptor: | 8-AZAXANTHINE, AZIDE ION, SODIUM ION, ... | | Authors: | Gabison, L, Colloc'H, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-10-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Azide and cyanide have different inhibition modes of urate oxidase
To be Published
|
|
5H66
 
 | |
5H67
 
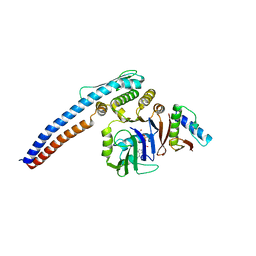 | |
5JUS
 
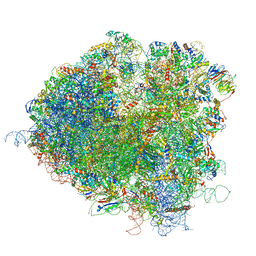 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure III (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5LH8
 
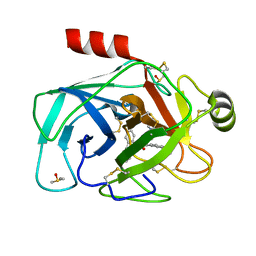 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 8 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)-3-methoxy-phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-[(1~{S},2~{R})-2-phenylcyclohexyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3OBP
 
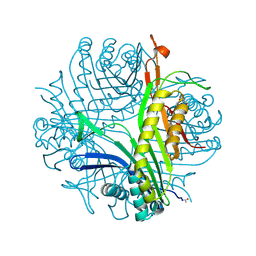 | | Anaerobic complex of urate oxidase with uric acid | | Descriptor: | SODIUM ION, URIC ACID, Uricase | | Authors: | Gabison, L, Chopard, C, Colloc'h, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-08-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray, ESR, and quantum mechanics studies unravel a spin well in the cofactor-less urate oxidase.
Proteins, 79, 2011
|
|
5JUP
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure II (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
4PUV
 
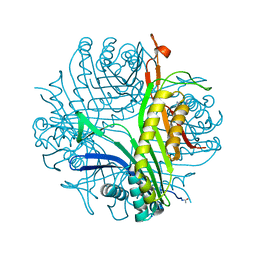 | | URATE OXIDASE DI-AZIDE complex | | Descriptor: | AZIDE ION, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Prange, T. | | Deposit date: | 2014-03-14 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Azide inhibition of urate oxidase.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5JUU
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure V (least rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5JUT
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure IV (almost non-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
4PR8
 
 | |
3P9O
 
 | | Aerobic ternary complex of urate oxidase with azide and chloride | | Descriptor: | AZIDE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gabison, L, Colloc'H, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Azide and Cyanide Show Different Inhibition Modes to Urate Oxidase
To be Published
|
|
3PK8
 
 | | Urate oxidase under 0.5 MPa / 5 bars pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
